Ribosomal RNA And Transfer RNA
Ribosomal RNA (rRNA)
Ribosomes are cell organelles, that function as a cell’s protein factories.
- They provide for the sites of protein synthesis involving the translation of the genetic information encoded in the messenger RNA (mRNA).
- When not engaged in protein synthesis, the ribosomes in a bacterial cell are always free, but in an eukaryotic cell, these may be found either free in the cytosol or attached to the membrane of ER (called rough endoplasmic reticulum).
- The initial progress in understanding the detailed structure of the ribosome came not from observing them with the electron microscope but by analyzing their components by ultracentrifugation.
- Ribosomes, as well as other small particles and molecules, are measured in units that describe their rate of sedimentation during density gradient centrifugation in sucrose.
- This technique gives information on size and shape (due to the speed of sedimentation) while simultaneously isolating the molecules.
- Isolation by centrifugation in sucrose is a relatively gentle isolation technique; the molecule still retains its biological properties and can be used for further experimentation.
- In the 1920s, physical chemist T. Svedberg developed ultracentrifugation, giving his name to the unit of sedimentation: the Svedberg unit, S.
- In sucrose density-gradient centrifugation, the gradient is formed by layering on decreasingly concentrated sucrose solutions.
- In a related technique, cesium chloride density-gradient centrifugation, the gradient develops during centrifugation.
The sucrose centrifugation is stopped after a fixed time, whereas in the cesium chloride technique, the system spins until it reaches equilibrium.
- The sucrose method tends to be more rapid. Samples can be isolated from a sucrose gradient by punching a hole in the bottom of the tube and collecting the drops in sequentially numbered containers.
- The first (lowest-numbered) containers will contain the heaviest molecules (with the highest S values).
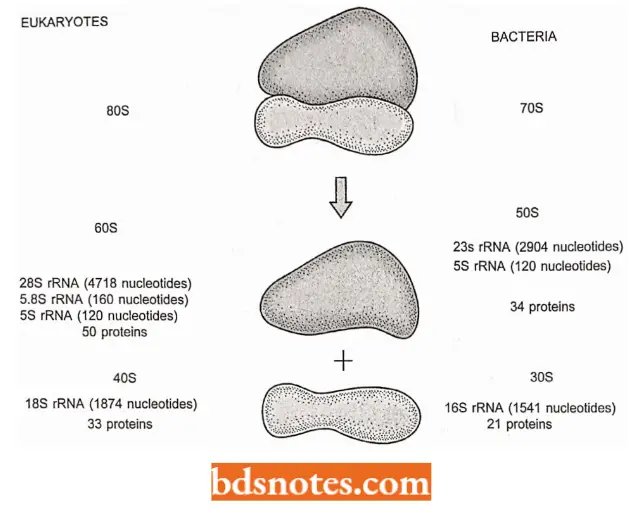
Components Of Ribosome: Intact ribosomes have sedimentation coefficients of 80S for eukaryotes and 70S for bacteria, and each can broken down into smaller components:
- Each ribosome comprises two subunits of unequal size. In eukaryotes these subunits are 60S and 40S, in bacteria they are 50S and 30S.
- Note that sedimentation coefficients are not additive because they depend on shape as well as mass; it is perfectly acceptable for the intact ribosome to have an S value less than the sum of its two subunits.
- The large subunit contains three rRNAs in eukaryotes (the 28S, 5.8S, and 5S rRNAs). In bacteria, the equivalent of the eukaryotic 5.8 rRNA is contained within the 23S rRNA.
- The small subunit contains a single rRNA in both types of organisms : an 18S rRNA in eukaryotes and a 16S rRNA in bacteria.
- Both subunits contain a variety of ribosomal proteins, the numbers.

- The ribosomal proteins of the small subunit are called SI, S2, S3, etc.; those of the large subunit are called L1, L2, etc. There is just one of each protein per ribosome, except for L7 and LI2, which are present as dimers.
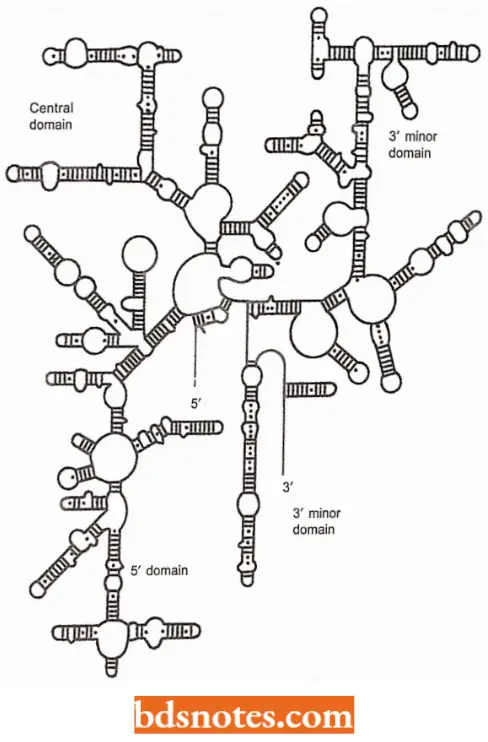
Investigation Of The Fine Structure of Ribosome: Once the basic composition of eukaryotic and bacterial ribosomes had been worked in water, attention was focused on how the various molecules of rRNAs and proteins fit together.
- Important information was provided by the first rRNA sequences, comparisons between these identifying conserved regions that can base-pair to form complex two-dimensional structures.
- This suggested that the rRNAs provide a scaffolding within the ribosome, to which the proteins are attached, an interpretation that under-emphasizes the active role that rRNAs play in protein synthesis.
Much of the subsequent research has concentrated on the bacterial ribosome, which is smaller than the eukaryotic version and available in large amounts from extracts of cells grown to high density in liquid cultures.
The Following Technical Approaches Have Been Used To Study The Bacterial Ribosome:
- Nuclease protection studies enable contacts between rRNAs and proteins to be identified.
- Protein-protein crosslinking identifies pairs or groups of proteins that are located close to one another in the ribosome.
- Electron microscopy has gradually become more sophisticated, enabling the overall structure of the ribosome to be resolved in greater detail.
- Innovations such as immunoelectron microscopy, in which ribosomes are labeled with antibodies specific for individual ribosomal proteins before examination, have been used to locate the positions of these proteins on the surface of the ribosome.
- Site-directed hydroxyl radical probing makes use of the ability of Fe ions to generate hydroxyl radicals that cleave RNA phosphodiester bonds located within any of the sites of radical production.
- This technique was used to determine the exact positioning of ribosomal protein S5 in the ribosome of E. coli.
- Different amino acids within S5 were labeled with Fe(II) and hydroxyl radicals induced in the reconstituted ribosomes. The positions at which 16S rRNA was cleaved were then used to infer the topology of the rRNA in the vicinity of the S5 protein (Heilek and Noller, 1996).
- X-ray crystallography. In recent years discussed techniques have been increasingly supplemented by X-ray crystallography which has been responsible for the most exciting insights into ribosome structure.

Analyzing the bulk amounts of X-ray diffraction data that are produced by crystals of an object as large as a ribosome is an enormous task, particularly at the level needed to obtain a structure that is detailed enough to be informative about how the ribosome works (Pennisi, 1999).
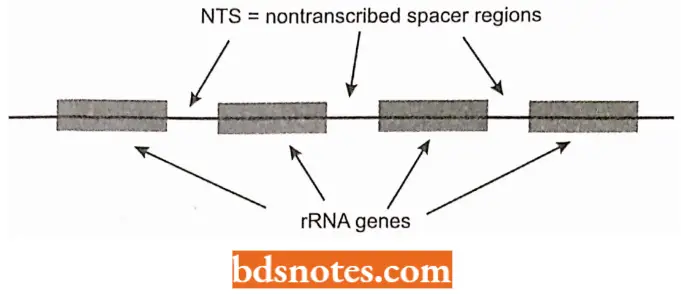
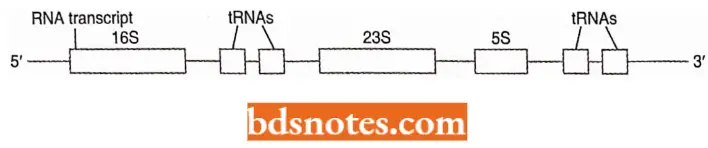
- In E. coli, all three ribosomal RNA segments are transcribed as a single long piece of RNA that is then cleaved and modified to form the final three pieces of RNA (i.e., 16S, 23S, and 5S).
- The region of DNA that contains the three ribosomal RNA molecules also contains genes for four tRNAs. There occur about ten copies of this region in each chromosome of E. coli.
- The occurrence of three r RNA segments on the same piece of RNA ensures a final ratio of 1:1:1, the ratio needed for ribosomal construction.
Nucleolus And Transcription In Eukaryotes: In eukaryotes, all rRNAs (i.e., 5.8S, 18S, and 28S) but the 5S ribosomal RNA section are transcribed as part of the same piece of RNA. However, eukaryotic cells have many copies of these rRNA genes, depending on the species.
- For example, the fruit fly, Drosophila melanogaster, has about 130 copies of the DNA region in the larger segments of ribosomal RNA that are transcribed.
- These regions occur in tandem on the X and Y sex chromosomes and are known collectively as the nucleolar organizer.
- The smallest ribosomal RNA (i.e., 5S rRNA) is also produced from a duplicated gene, but at a different point in the genome.
- For example, in D. melanogaster, the 5S rRNA is produced on chromosome It is still not known why 5S rRNA is transcribed separately.
- As we know eukaryotes have three RNA polymerases (enzymes).
- Eukaryotic RNA polymerase 1 (or polymerase A) transcribes only the nucleolar organizer DNA (i.e., most rRNAs except 5S).
- RNA polymerase 2 (or polymerase B) transcribes most genes (i.e., genes for mRNAs).
- RNA polymerase 3 (or polymerase C) transcribes small genes, primarily the 5S rRNA gene and tRNA genes.
- Primase does primer synthesis during DNA replication. In addition, mitochondria, chloroplasts, and some phages have other RNA polymerases.
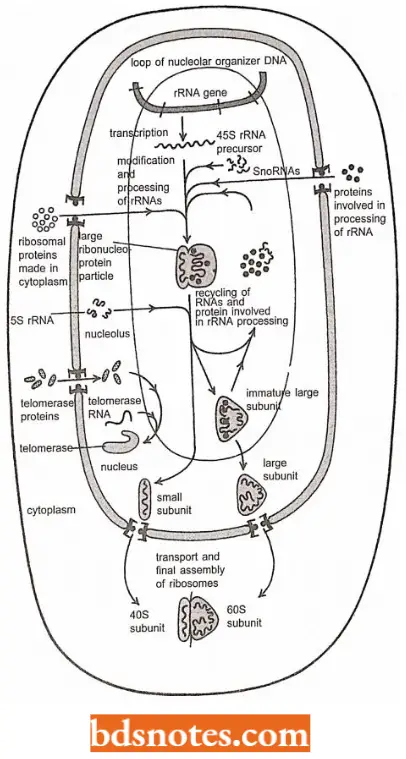
- At the nucleolar organizer, the nucleolus forms a dark globule found in eukaryotic nuclei.
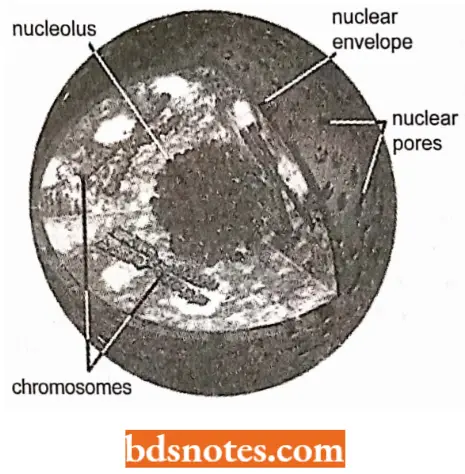
- The nucleolus is the place where ribosomes are assembled.
- The various ribosomal proteins that have been synthesized in the cytoplasm migrate to the nucleus and eventually to the nucleolus, where, with the final forms of the rRNA, they are assembled into ribosomes.
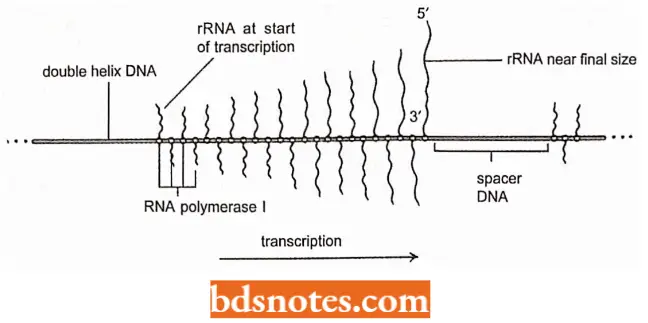
- In the nucleolar organizer, an untranscribed region of spacer DNA separates each repeat of the large ribosomal DNA gene.

- In the electron micrograph, the polarity of transcription is evident from the short RNA at one end of the transcribing segment and the long RNA at the other ends, with a uniform gradation between.
- Notice that many RNA polymerases are transcribing each region at the same time.
- The regions between the transcribed DNA segments are the spacer DNA regions.
- Because each RNA gene has a fixed initiation site (promoter) and a fixed termination site, the transcripts adopt the characteristic Christmas tree configuration.
Processing of Ribosomal RNAs: Three of four rRNAs (185, 5.8, and 28S ) are made by chemically modifying and cleaving a single large (45S) precursor rRNA.
- Extensive chemical modifications occur in the 13,00-nucleotide- long precursor rRNA before the rRNAs are cleaved out of it and assembled into ribosomes. (Note. The 5SrRNA does not require chemical modification).
- Such modifications include about 10 methylations of the 2’-OH positions on nucleotide sugars and 100 isomerizations of uridine nucleotides to pseudouridine.
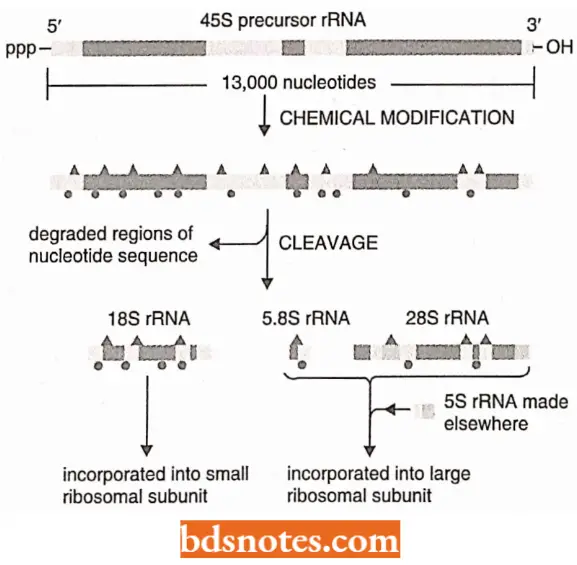
- The functions of these modifications are not understood in detail, but they probably aid in the folding and assembly of the final rRNAs and may also delicately alter the function of ribosomes.
- Each modification is made at a specific position in the precursor RNA.
- These positions are specified by several hundred “guide RNAs”, which locate themselves through base pairing to the precursor rRNA and thereby bring an RNA modifying enzyme to the appropriate position.
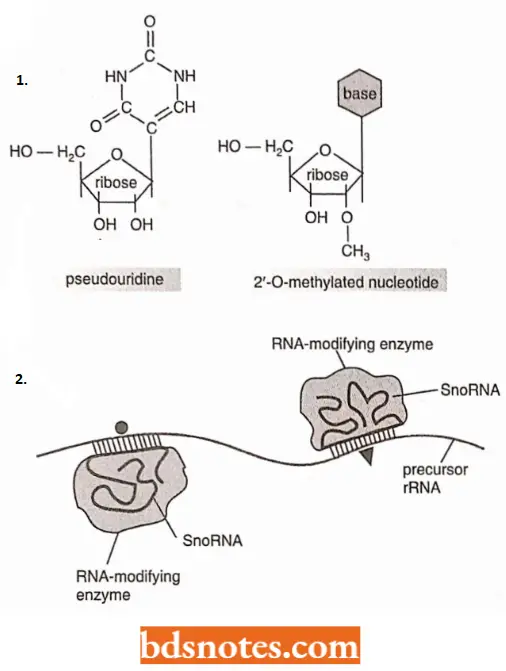
- Other guide RNAs promote, probably by causing conformational changes in the precursor RNA, the cleavage of the precursor rRNA into the rRNAs, All of these guide RNAs are members of a large class of RNA, called small nucleolar UN As (or snoRNAs).
- So named because these RNAs perform their functions in a sub-compartment of the nucleus, the nucleolus.
- Many snoRNAs are encoded in the introns of other genes especially those encoding ribosomal proteins.
- They are therefore synthesized by RNA polymerase 2 and processed from excised intron sequence.
Role Of Nucleolus In Transcription: In addition to the important role in ribosome biogenesis, the nucleolus is also the site where other RNAs are produced and other RNA protein complexes are assembled.
- For example, the U6 snRNP which functions in pre-mRNA splicing, comprises one RNA molecule and at least seven proteins. The U6 snRNA is chemically modified by snoRNAs in the nucleolus before its final assembly there into the U6 snRNP.
- Other important RNA protein complexes including telomerase and the signal recognition particle, are also believed to be assembled at the nucleolus.
- Finally, the tRNAs are processed as well in the nucleolus.
- Thus the nucleolus can be thought of as a large factory at which many different noncoding RNAs are processed and assembled with proteins to form a large variety of ribonucleoprotein complexes.
Ribosomal RNA Multiple Choice Questions And Answer
Question 1. The function of nucleolus is the synthesis of
- DNA
- mRNA
- rRNA
- tRNA
Answer: 3. rRNA
Question 2. The functional unit in the synthesis of protein is
- Peroxisome
- Dictyosome
- Lysosome
- Polysome
Answer: 4. Polysome
Question 3. An anticodon of tRNA represents
- Wobble hypothesis
- Template hypothesis
- Gene flow hypothesis
- Richmond and Long effect
Answer: 1. Wobble hypothesis
Question 4. Which of the following RNAs picks up specific amino acids from the amino acid pool in the cytoplasm to the ribosome during protein synthesis?
- tRNA
- mRNA
- rRNA
- All of them
Answer: 1. tRNA
Question 5. Which site of the tRNA molecule hydrogen bonds to a mRNA molecule?
- Codon
- Anticodon
- 5’end of tRNA
- 3’end of tRNA
Answer: 2. Anticodon
Question 6. The cloverleaf model belongs to
- tRNA
- DNA
- Centriole
- Flagella
Answer: 1. tRNA
Question 7. An anticodon is complementary to the nucleotide triplet in
- tRNA
- rRNA
- mRNA
- cDNA
Answer: 3. mRNA
Question 8. Which of the characters does not apply to tRNA?
- It is the smallest of the RNAs
- It acts as an adapter for amino acids
- It has a clover leaf structure
- It is the largest of the RNAs
- It bears anticodon
Answer: 4. It is the largest of the RNAs
Question 9. According to Wobble’s hypothesis
- The first base is unstable
- The second base is unstable
- The third base is unstable
- The process of polypeptide chain elongation has been established
Answer: 3. Third base is unstable
Question 10. The codon for anticodon 3’UUA 5′ is
- 5’AAU 3′
- 3’AAU5′
- 5’AAT 3′
- 3′ AAC5′
Answer: 1. 5’AAU 3′
Question 11. tRNA recognizes aminoacyl synthetase enzymes by
- Anticodon
- DHU loop
- T C loop
- AA-site
Answer: 2. DHU loop
Question 12. Shape of 3 – D view of tRNA is
- Z- shaped
- X-shaped
- Y-shaped
- L-shaped
Answer: 4. L-shaped
Question 13. The amino acid binding site in tRNA is
- 5′ end
- Anticodon loop
- CCA 3’end
- DHU loop
Answer: 3. CCA 3’end
Question 14. Genetic information for the synthesis of ribosomal RNA is coded in
- DNA present in the nucleus
- Nucleolar associated chromatin
- Granular zone of nucleolus
- Amorphous zone of nucleolus
Answer: 2. Nucleolar associated chromatin

Leave a Reply