Multigene Families
The majority of genes are spaced out more or less randomly along the length of the DNA molecule. In some cases, however, they are grouped into distinct clusters.
- Sometimes the individual genes in a cluster are unrelated and there is no apparent reason or advantage to having them organized in this way.
- More commonly, clusters are made of genes that contain related units of biological information, for example., operons and multigene families.
Operons
Operons are quite common aspects of the organization of genes in bacteria. An operon is a cluster of genes coding for a series of enzymes that work in concert in an integrated biochemical pathway.
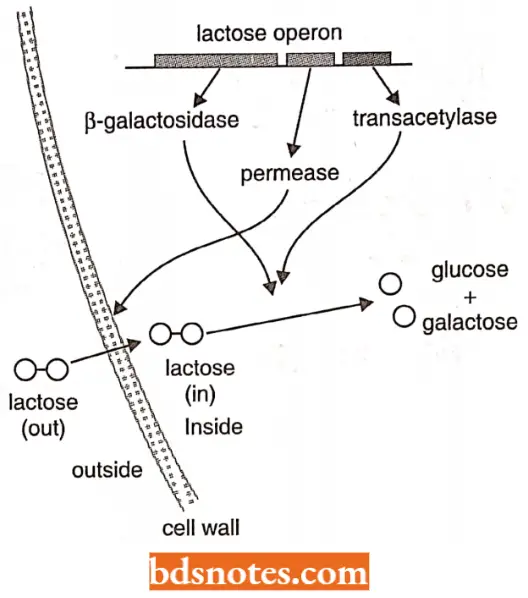
- The first operon to be discovered was the lactose or lac operon of E.coli which is a cluster of three genes each coding for one of the three enzymes involved in the conversion (catabolism) of the disaccharide lactose into its monosaccharide units glucose and galactose.
- These enzymes are not required all the time, only when lactose is available.
- But when needed they are required together, for example., the three units of biological information carried by Lac must be read at the same time.
- A sophisticated system enabling the lac genes to be expressed together and only when needed has evolved around the fact that the genes are clustered.
Multigene Families
Operons have no direct counterparts in organisms other than bacteria. In contrast, multigene families are found in many organisms, mainly eukaryotic organisms.
- A multigene family or gene family is also a cluster of related genes.
- Still, in multigene families, the individual genes have identical or similar nucleotide sequences and therefore contain identical or similar pieces of biological information.
Why do IViultigene Families are Required?
Some proteins in the cell are required in small quantities, but other proteins may be required in large quantities so that the demands can not be easily met. These demands can be met in either of the following two ways:
- In some cases, a solitary gene is present in the cell but is transcribed repeatedly to produce a large number of messenger RNA molecules, and these mRNA molecules act as templates for many rounds of protein synthesis.
- For example, although the gene coding for silkworm protein fibroin is present in only one copy per haploid complement, it keeps on making silk actively.
- Similarly, in a cell (for example., erythroblasts) engaged in exclusive synthesis of hemoglobin, a solitary copy of the gene produces large quantities of a and (3 polypeptide chains of hemoglobin.
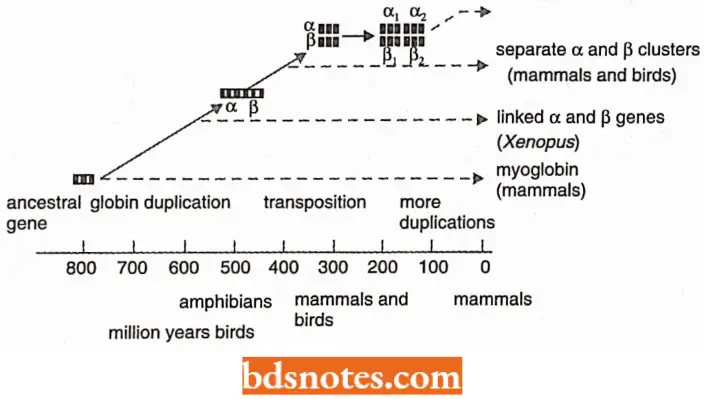
- However, although a whole family of related globin genes are found in each cell, different but solitary globin genes function at different stages of development.
- In other cases, multiple copies of a gene are found in the same cell, all taking part in transcription within a cell, giving rise to large quantities of the gene product as in the case of ribosomal RNA (rRNA) genes, histone genes, tRNA genes, heat shock protein genes, etc.
Types Of Multigenes: Members of multigene families share DNA sequence homology, descend from a single ancestral gene, and their gene products frequently have similar functions. Members of multigene families are often, but not always, found together in a single location along a chromosome.
Dispersed Family Of Gene: In some multigene families, the individual members are clustered, as with the globin genes, but in others, the genes are dispersed around the genome.
- An example of a dispersed family is the five human genes for aldolase, an enzyme involved in energy generation, which is located on chromosomes 3, 9,10, 16, and 17.
- The important point is that, even though dispersed, the members of the multigene family have sequence similarities that point to a common evolutionary origin.
Multigene Families Are Of The Following Main Types:
- Simple Multigene Families:
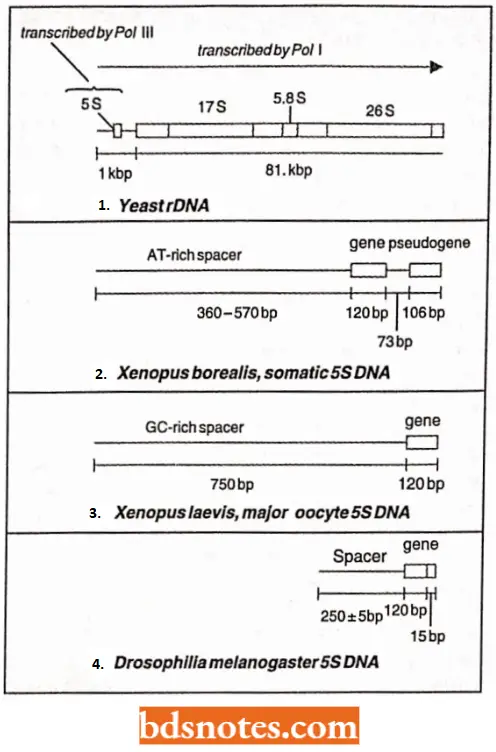
- In these gene families, all of the genes are exactly or virtually the same. For example, most higher organisms have multiple copies of the gene for the 5S rRNA (a component of the ribosome), probably because large quantities of 5S rRNA molecules must be synthesized at certain times, creating a demand that just one of a few genes would be unable to satisfy.
- Rather than being spread randomly throughout the chromosomes, the 5S rRNA genes are clustered into a multigene family. In humans, for example, there is a family of about two thousand 5S rRNA genes in chromosome 1.
- The 5S rRNA genes are present independently of the rDNA which is localized at the NOR in eukaryotes. However, in prokaryotes and yeast, 5S rRNA genes are present in close vicinity of rDNA. The 5S rRNA genes are also organized in tandem repeats, each repeat consisting of a gene 120 bp long and a spacer region. The length of complete repeat is 375 bp in Drosophila.
- In wheat, there are two loci for 5S rRNA having repeats of different lengths, 480 bp and 500 bp. These repeat units may sometimes carry pseudogenes (instead of functional genes), which are not used for the synthesis of any RNA.
- Complex Multigene Families:
- A multigene family is a group of similar, but not necessarily identical sequences, each sequence representing a gene, so that the gene is present in multiple copies. Complex multigene families are of the following types:
Multigene Families With Divergent Members
Globin Gene Family In The Human Genome: The human α-globin and β-globin genes are subfamilies of the globin gene superfamily and are two of the most intensively studied regions of the human genome.
- There is a cluster of three α-globin β-genes on the short arm of chromosome 16 and another with five β- globin genes on the short arm of chromosome 11.
- Members of both subfamilies share nucleotide sequence similarity, but members of the same subfamily have the greatest sequence similarity.
Haemoglobin is a tetramer, containing twoα- and two β- polypeptides. Each polypeptide incorporates a haem group that reversibly binds oxygen.
- Within each subfamily, genes are coordinately turned on and turned off during embryonic, fetal, and adult stages of development.
- For both the alpha and beta subfamilies, this expression occurs in the same order in which the genes are arranged on the chromosome.
- The alpha subfamily spans more than 30 kb and contains three genes: the (zeta) gene, expressed only in the early embryonic stage, and two copies of the gene, expressed during the fetal (α1) and adult stages (α2).
- In addition, two nonfunctional pseudogenes (ψ z and ψ) are present in the cluster. Pseudogenes are designated by the prefix ψ (psi), followed by the symbol of the gene that they most resemble. Thus, the designation ψ α1 indicates a pseudogene of the adult α, gene.
- Pseudogenes are non-functional versions of genes that resemble other gene sequences but contain significant nucleotide substitutions, deletions, and duplications that prevent their expression.
- The organization of the alpha subfamily members and the location of their introns and exons reveal several interesting features.
- First, as is common in eukaryotes, the DNA encoding the three functional genes occupies only a small portion of the region containing the subfamily. Most of the DNA in this region is intergenic spacer.
- Second, each functional gene in this subfamily contains two introns at precisely the same positions.
- Third, the nucleotide sequences within corresponding exons are nearly identical in the z and genes.
- Both genes encode polypeptide chains of 141 amino acids. However, their intron sequences are highly divergent, even though they are about the same size. Significantly, much of the nucleotide sequence of each gene is contained in these noncoding introns.

- The human β- globin gene cluster is longer than the α- globin cluster and contains five genes spaced over 60 kb of DNA. As with the alpha subfamily, the order of genes on the chromosome parallels their order of expression during development.
- Of the five genes, three are expressed before birth. The ε(epsilon) gene is expressed only during embryogenesis, while the two nearly identical γ(gamma) genes (Gγ and Aγ) are expressed only during fetal development.
- The polypeptide products of the two y-genes differ only by a single amino acid. The two remaining genes, δ(delta) and β(beta), are expressed following birth. Finally, a single pseudogene ψβ1 is present within the subfamily.
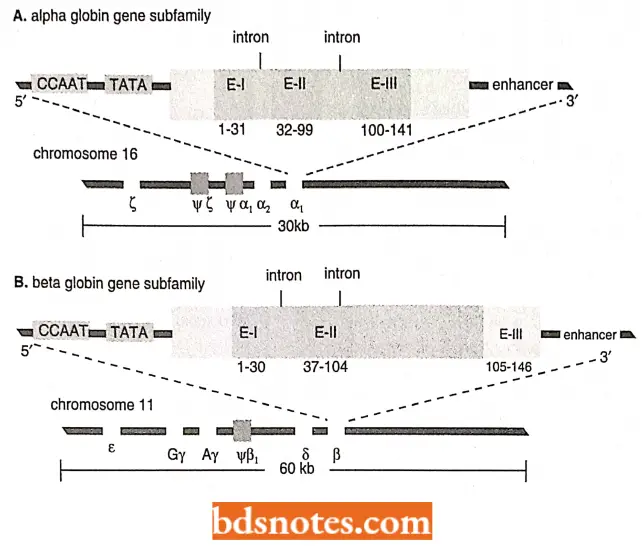
All five functional genes encode proteins with 146 amino acids and have two similarly sized introns at the same positions. The second intron in the P gene is significantly larger than its counterpart in the functional a genes.
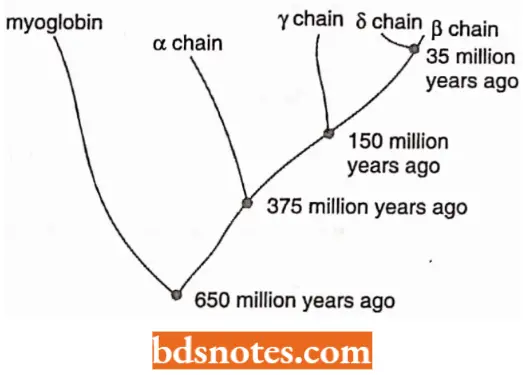
- These similarities reflect the evolutionary history of each subfamily and the events such as gene duplication, nucleotide substitution, and chromosome translocations that produced the present-day globin superfamily.
- It has been shown that in these sequences (cluster genes and p cluster genes), the coding regions show little divergence, but spacers in between the genes show considerable diversity as in the case in many other gene families.
- The only explanation for this is that the protein function puts a constraint on the evolution of the coding sequence, while no such constraint is exercised in the spacer region which is fast evolving. Based on the degree of similarity in amino acid sequences of globin of different mammals, evolutionary trees have been prepared in the past.
Actin Gene Family In Eukaryotes: Actin is a cytoskeleton protein. Actin genes represent a multigene family, where the members are homologous, but non-identical, giving rise to slightly different variants so that different members function either at different times in the same tissue or in different tissues at the same time.
For example, a minimum of six closely related polypeptides are synthesized in different cells at different times in mammals; the number may be larger in some flowering plants. The actions may be broadly classified as a,(3 and y chains.
- Of these, actins make the contractile apparatus of the muscle, while [3 and y actins are found together in almost all nonmuscle cells.
- There may be variants in each of these three classes, making up to 20-30 actin-related sequences in the human genome; humans have six genes for actin.
- In several plant systems (for example., tobacco, potato, etc.) also, more than 20 related but variant actin sequences have been reported.
- Actin is extraordinarily well-conserved among eukaryotes. The amino acid sequence of actions from different species is usually about 90% identical. Yeast actin and Drosophila muscle actin are 89% identical.
Other multigene families with divergent members: Other multigene families have divergent members; for example.,
- Albumin, a fetoprotein (major component of blood plasma), serine proteases, the interferons (for defense against a viral infection), and the immunoglobulins;
- Chorion proteins make the major components of egg shells in insects.
Multigene families with identical genes: In some cases, for producing large quantities of a gene product, many copies of identical genes are required. These genes may occur as repeat units, each of which comprises
- A coding region, which is highly conserved to produce the same gene product and
- A spacer region may show divergence. Examples of these multigene families are the following
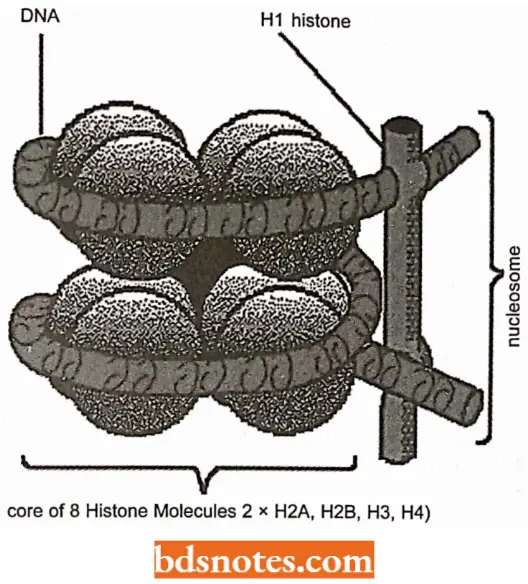
Multiple copies of histone genes. The organization of the histone genes is a variation on the theme established in the globin gene family.
- Here a cluster of five related, but nonidentical genes, separated from each other by highly divergent intergenic spacer regions, is tandemly repeated many times.
- As we already know histones are positively charged (basic) proteins which interact with the negatively charged phosphate groups of DNA to form nucleosomes, the chromatin units.
- In rapidly diving cells (for example., a cleaving fertilized egg of a frog), histone synthesis must keep pace with DNA replication, and the necessary quantity of histones must appear each time, as one cell cycle runs into the next.
- Multiple copies of the histone Histones, cluster allow the coordinate synthesis of DNA and histone during the S phase of the cell cycle. Many interesting correlations support this idea.
- Yeast cells, with much less DNA than other eukaryotes, have only two clusters of four histone genes(they lack histone HI).
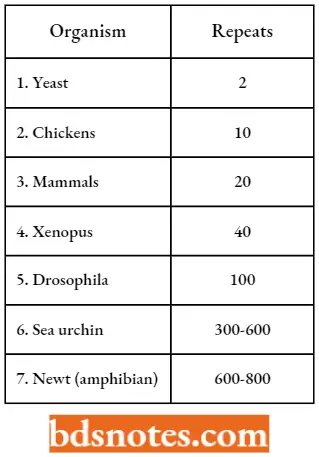
- In contrast, clusters of five histone genes are tandemly repeated from 10 to 800 times in many complex organisms. Mitotic cell division is very rapid during development in sea urchins and newts and these organisms have one of the largest set of histone gene clusters.
Numbers Of Histone Gene Clusters In Selected Eukaryotes:
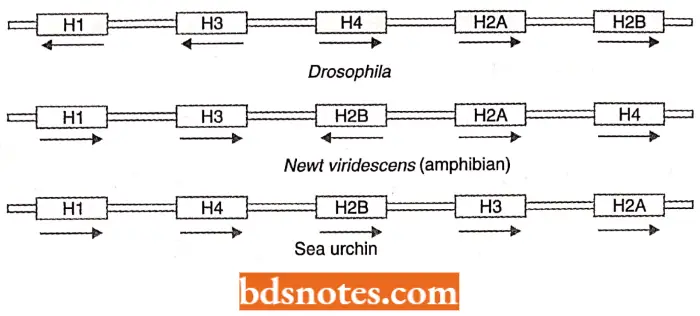
Polarity differences in transcription: Beyond its tandem arrangement, the histone gene family shows several other differences from other families.
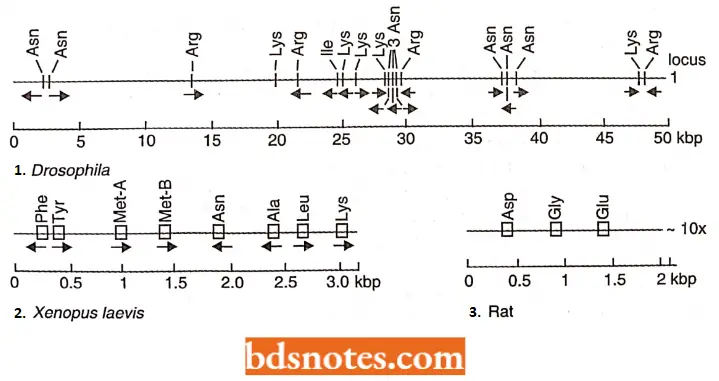
- First, almost all histone genes lack introns. Second, individual genes within clusters of a given species often orient in opposite directions concerning transcription.
- As shown, the arrangement of genes and their polarity of transcription, shown by the direction of the arrow, vary in the sea urchin and Drosophila, two organisms where this cluster has been extensively studied. Studies also show polarity differences in other organisms.
- Despite polarity differences, the amino acid sequences of the various histones (reflecting the DNA encoding each one) are remarkably similar in highly divergent organisms. The homology of histone genes is one of the best examples of sequence conservation through evolution.
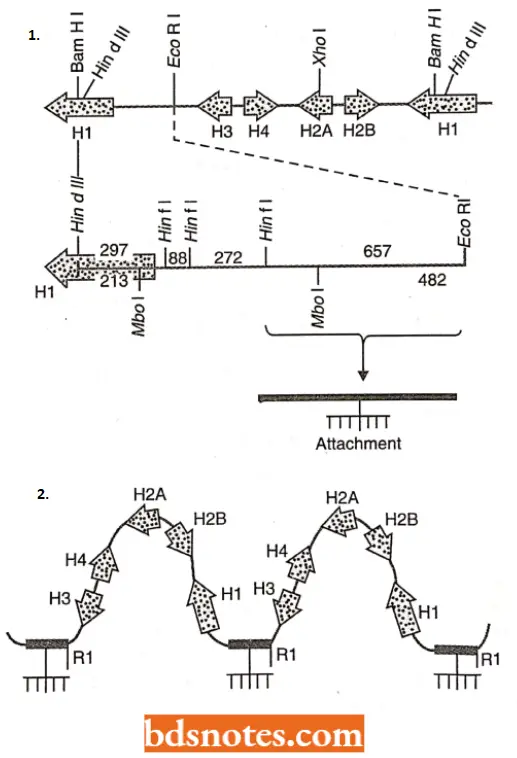
- Another interesting feature of the multigene family of histone genes observed in Drosophila is the presence of an ‘ attachment site’1 per repeat unit; 100 such repeat units being present in each haploid genome.
- With the help of the attachment site, the genes (DNA) remain attached to the nuclear scaffold. These attachment sites help in transcription.
- In mice and humans, members of the histone gene family are clustered but do not form tandem arrays. A region at the distal tip of the long arm of human chromosome 7 contains all copies of the histone gene family, interspersed with other nonhistone genes.
- Therefore, no single pattern of histone gene organization applies to all organisms.
- Ribosomal RNA (rRNA) genes in tandem arrays. For the production of about 10 million ribosomes per eukaryotic cell, rRNA genes are present in multiple copies at nucleolar organizing regions(NOR) of specific satellited chromosomes.
The number of these genes may vary from 50 to 30,00 in a cell and this number may be unequally distributed on NORs if more than one such loci are present.
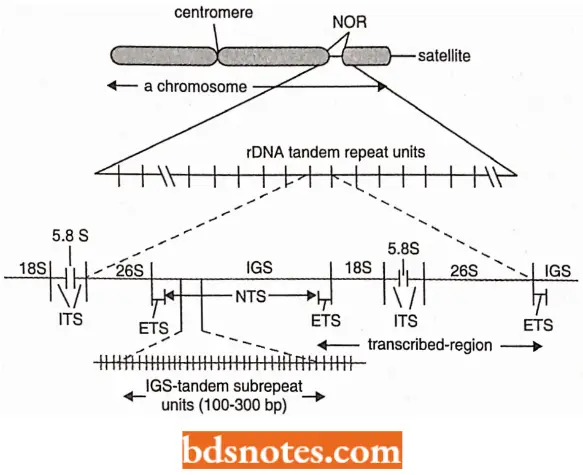
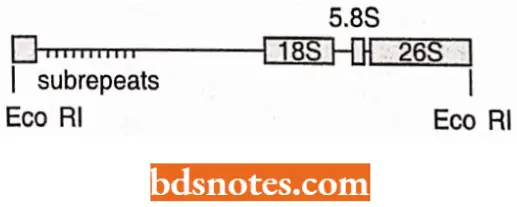
The DNA comprising these genes is called rI)NA (ribosomal DNA), which is repetitive. Each repeat unit has
- A coding region with genes that specify 18S, 5.8S and 28S rRNA molecules;
- A spacer region called intergenic spacer (IGS) and
- Internal transcribed spacers (ITS) one each between 18S and 5.8S genes and between 5.8S and 28S genes. Because parts of the intergenic spacer (IGS) adjoining region, known as an external transcribed spacer (ETS), is also transcribed, the use of the term non-transcribed spacer (NTS) for the whole spacer region has been considered to be inappropriate. The NTS makes only a part of IGS, the remaining part of IGS being ETS.
The IGS region in its turn has a region consisting of a tandem array of variable number of subrepeats ranging from 100 – 300 bp (bp = base pairs) in length. The variation in the number and size of the sub-repeats in IGS is responsible for the variation in the length of the repeat units (IGS + coding region).
The rRNA repeat units are usually looped off from the main chromosome fibers, in the form of extended threads at the nucleolar organizing region. These loops, in association with specific proteins, form the nucleoli, where rRNA synthesis and processing take place.
- The number of nucleoli and the corresponding number of nucleolar organizing chromosomes in an organism may vary from one to several.
- At each locus (NOR), rDNA repeat units may evolve independently both in length and also in the nucleotide sequence of the spacer region.
- As a result of this, the length of the repeat unit including coding and spacer regions (non-transcribed spacer or NTS; external transcribed spacer or ETS and internal transcribed spacers or ITS) varies from about 7 kb (kilobase) pairs to 14 kb. In wheat and barley, it is usually 9 kb to 10 kb.
Transfer RNA (tRNA) Mulligan Families: The genes for each of the different tRNAs are also found in multiple copies to meet the heavy demands of the cell for the production of tRNA molecules. Ten to several hundred genes for each IRNA are present in each haploid genome.
- While in some cases with fewer copies of a gene, these copies are dispersed, in other cases such as in Xenopus (a load, an amphibian), tandem repeats of long sequences (each repeat having genes for several different tRNAs) are found.
- In Xenopus, for example, a 3.2 kb repeat length has 8 genes, 2 for tRNA and six others, for six different 3′ tRNA molecules.
- In still other cases (for example., Drosophila) genes for completely different tRNA species arc clustered over a length of several thousand base pairs. Sometimes, tRNA pseudogenes are also found.
- Small nuclear RNA (Sn RNA) genes. Small nuclear RNAs (snRNAs) are found in abundance in the nuclei of all eukaryotes and represent neither IRNA nor rRNA.
- Six snRNAs, which are usually found, range from 100 (U6) to 215 (U3) nucleotides in length and are involved in RNA processing through the formation of snRNP (small nuclear ribonucleoproteins).
- SnRNAs are encoded by multiple copies of identical genes organized in tandem arrays of repeat units, each SnRNA gene being flanked on either side of spacer DNA ranging from 800 bp to 45,000 bp in length in different organisms.
- DNA of genes of Sn RNAs is reported to have a higher ratio( 10:1) of pseudogenes than functional genes. In humans, for example, there are 30 genes, for U1 (an SnRNA) on chromosome 1 and 500 to 1000 pseudogenes distributed throughout the genome.
- Multigene families for storage proteins in crop plants. Storage proteins of several crop plants have been reported to be encoded by multigene families and are represented mainly as prolamins ( major storage proteins in cereals; soluble in aqueous alcohol) and oxglobulins (major storage proteins in legumes; soluble in salt).
- Three to ten genes for each of the ten prolamin families in wheat; and 18 genes belonging to three families of globulin genes are known in pea.
- Homologies in genes coding for prolamins and globulins in different crop plants have been reported. Their multiplicity appropriately meets the demand for rapid synthesis of these proteins in the developing seeds.
Concerted Evolution Of Multigene Families
Multigene families are found to have mechanisms that prevent the individual copies from accumulating mutations and hence diverging away from the functional sequence. This is called concerted evolution.
Thus, if one copy of the family acquires an advantageous mutation then that mutation can spread throughout the family until all members possess it. Following genetic mechanisms may be involved in such homogenization of concerted evolution:
- Gene conversion: Gene conversion can result in the sequence of one copy of a gene being replaced with all or part of the sequences of a second copy. Multiple gene conversion events could therefore maintain identity among the sequences of the individual members of a multigene family.
- Unequal crossing over Unequal crossing over may lead to duplications or deletions leading to homogenization, i.e., either mutant member will be deleted or multiply and spread throughout the gene family. In yeast, it has been reported that the rates of gene conversion and unequal crossing-over are higher than the rate of mutations so that homogeneity can be maintained.
Further, certain specific sequences are also located in the multiple gene families which stimulate higher rates of recombination in a multigene family. In rDNA, the NTS (non-transcribed spacer) region is a hot spot of recombination thus facilitating homogenization.

Leave a Reply