DNA Replication
As a carrier of genetic information DNA has the following two important functions:
- Heterocatalytic function – When DNA directs the synthesis of chemical molecules other than itself (for example., synthesis of RNA, proteins, etc.), then such functions of DNA are called heterocatalytic functions.
- Autocatalytic functions – The functions of DNA which directs the synthesis of DNA itself, are called autocatalytic functions. Here, we are concerned only with the autocatalytic function of DNA.
Watson And Crick’s Model For DNA Replication
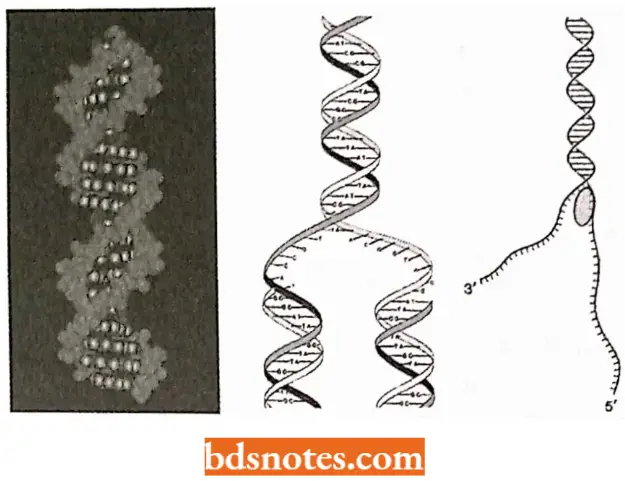
The double helix model of the DNA molecule of Watson and Crick beautifully embodied a built-in template system for self-replication or autocatalytic function.
- Because of the specificity of base pairing, the sequence of bases along one chain automatically determines the base sequence along the other.
- Thus, each chain of the double helix can serve as a template for the synthesis of the other. For the replication of DNA molecules.
- Watson and Crick proposed that replication involved the disruption of hydrogen bonds followed by a rotation and separation of the two polynucleotide strands.
- Each purine and pyrimidine base of each polynucleotide strand is thought to attract a complementary free nucleotide available for polymerization in the cell and to hold it in place using the specific hydrogen bonds.
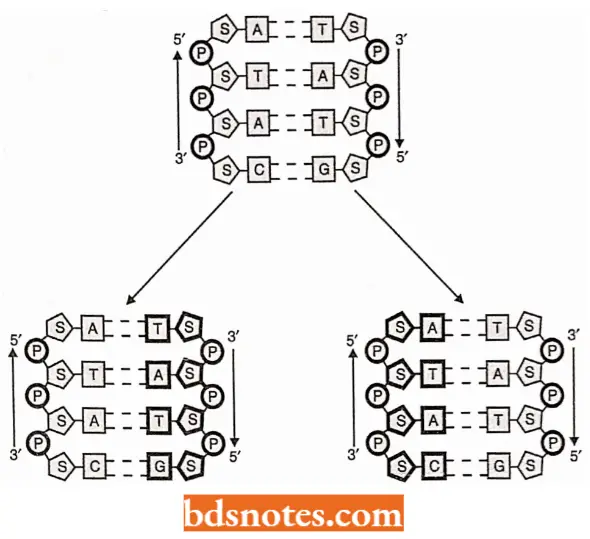
Once held in place on the parent template chain, the free nucleotides were sewn together by the formation of the phosphate diester bonds that linked adjacent deoxyribose residues, forming a new polynucleotide molecule of a predetermined base sequence. Thus, two double-helical molecules, identical to each other, are formed.
Experimental Evidence for Semiconservative DNA Replication in E.coli: The Watson – Crick’s model of DNA structure and replication suggests that once DNA replication is initiated, the two original polynucleotide strands of the duplex of helix will unwind, at least locally, so that each can serve as a template for a new strand.
- An immediate prediction follows from this proposal: both duplexes that result from replication should be hybrid, each containing an old strand derived from the original molecule and a new strand that has been formed during the replication process.
- Since each of the two double helices or duplexes conserves only one of the parent polynucleotide strands, the process is said to be semiconservative. This prediction is diagrammed in which old DNA is shown in black and new DNA in white.
- Also outlines what would be predicted if these two hybrid duplexes went on to replicate themselves. Four duplexes would result, two of which would contain a single strand derived from the original chromosome and two of which would contain new DNA strands.
- Besides the semiconservative mode of DNA replication, the following two methods of DNA replication were deemed equally feasible:
- Conservative replication, in which both strands of parent double helix would be conserved and the new DNA molecule would consist of two newly synthesized strands; and
- Dispersive replication, in which replication would involve fragmentation of the parent double helix and the intermixing of pieces of the parent strands with newly synthesized pieces, thereby forming the two new double helices.
![]()
Meselson and Stahl’s experiment: M. Meselson and F.W. Stahl (1958) verified the semiconservative nature of DNA replication in a series of elegant experiments using isotopically labeled DNA and a form of isopycnic density gradient centrifugation.
- They cultured Escherichia coli cells in a medium in which the nitrogen was 15N (a ‘heavy’ isotope of nitrogen, but not a radioisotope) instead of commonly occurring and lighter 14N.
- In time, the purines and pyrimidines of DNA in new cells contained 15N (where 14N normally occurs) and, thus, the DNA molecules were denser.
- DNA in which the nitrogen atoms are heavy (15N) can be distinguished from DNA containing light nitrogen (14N) because during isopycnic centrifugation, the two different DNAs band at different density positions in the centrifuge tube.
- Depending on its content of 15N and 14N, the DNA bands are at a specific position in the density gradient. Because the DNA synthesized by E.coli cells grown in,15N would be denser than 14N containing DNA, it would band further down the tube.
- E.coli cells’ growth for some time in the presence of 15N-medium was washed free of the medium and transferred to a 14N-containing medium and allowed to continue to grow for specific lengths of time(i.e., for various numbers of generation time).
![]()
- DNA isolated from cells grown for one generation of time in the 14N medium had a density intermediate to that of the DNA from cells grown only in the 15N-containing medium (identified as generation; and that of DNA from cells grown only in the 14N-containing medium (the controls).
- Such a result immediately ruled out the possibility that DNA replication was conservative because the conservative replication would have yielded two DNA bands in the density gradient for generation (i.e., F, cells).
- The single band of intermediate density (identified as “hybrid DNA”) consisted of DNA molecules in which one strand contained 15N and the other contained 14N.
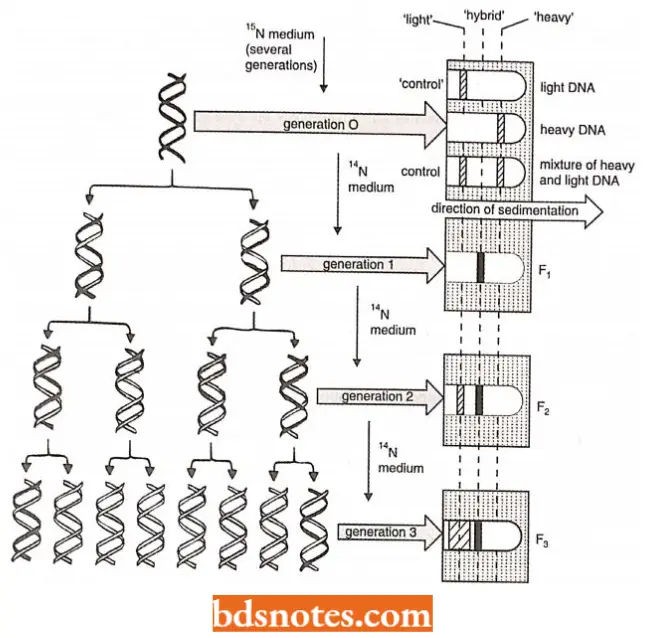
- When the incubation in the 14N-medium was carried out for two generations of time (for example., generation 2), two DNA bands were formed – one at the same density position as the DNA from cells grown exclusively in 14N medium (i.e., light controls) and the other of intermediate density.
- Subsequent generations produced greater numbers of DNA molecules that banded at the “light” (14N-containing DNA) position in the density gradient.
- These results are consistent only with the model of semiconservative replication. Studies using other prokaryotes as well as eukaryotes indicate that semiconservative replication of DNA is probably a universal mechanism.
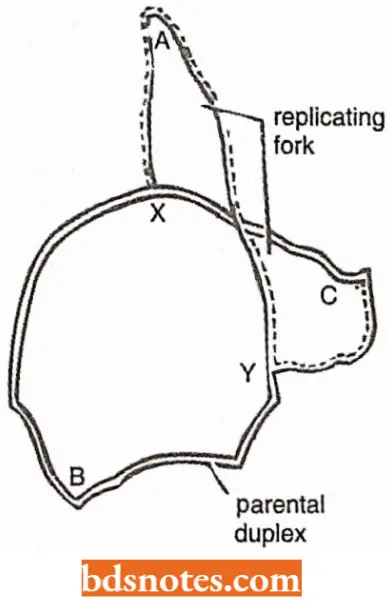
Visualization of replication in E.coli: In 1963, J.Cairns developed a technique employing a combination of microscopy and autoradiography that made it possible to visualize the replication of the chromosome of E.coli.
- Cairns placed E.coli cells in a medium containing 3H-thymidine (tritiated-thymidine)for various periods so that the radioactive thymidine was incorporated into the DNA as the chromosome was replicated in successive generations of cells.
- E. coli cells were removed from the medium after various periods of incubation and gently lysed to release the chromosome from the cell (since the shear forces created by harsh lysis break the chromosome into small pieces).
- The chromosomes were then transferred to glass slides and coated with a photographic emulsion sensitive to the low-energy beta particles emitted by the 3H-thymidine.
- After exposing the emulsion to the beta rays, the emulsion was developed and examined by light microscopy.
- Wherever decay of labeled thymidine had occurred in a chromosome, the emulsion was exposed and created visible grains. A chromosome not engaged in replication appeared as a circular structure formed from a close succession of exposure spots.
- Chromosomes “caught in the act” of replication gave rise to what is called theta configurations because they have the appearance of the Greek letter theta. The theta structures reveal the positions of the replication forks in the circular chromosomes.
- Cairns’ observations also clearly supported the semiconservative nature of replication. The rate at which the replication proceeds could also be worked out by measuring the length of DNA undergoing replication in a known interval of time.
- Cairns worked out the generation time of E.coli as 30 minutes. The length of the chromosome was worked out to about 1 mm. The rate of replication, thus, would be approximately 30 μm – 40 μm per minute (1mm = 1000 μm).
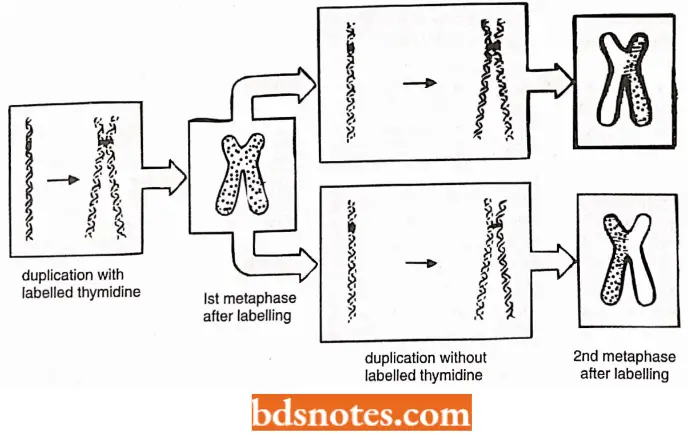
Evidence for Semiconservative Replication of Chromosomes (or DNA) in Eukaryotes: J.H. Taylor and P. Woods (1957) provided evidence in support of semiconservative mode of DNA replication in eukaryotes by using the technique of autoradiography and light microscopy in dividing root tip cells of the bean, Vicia faba.
- After incorporation of tritiated thymidine, when root tips were transferred to the unlabelled culture medium (and colchicine was added to the medium to prevent anaphase separation of sister chromatids).
- In the first generation of duplication both chromatids were labeled (this is interpreted as one DNA double helix in each chromatid and only one of the two strands labeled).
- In the second cycle of duplication (in the unlabelled medium) in each chromosome, one of the two chromatids was found to be labeled. This was interpreted as showing semiconservative mode duplication.
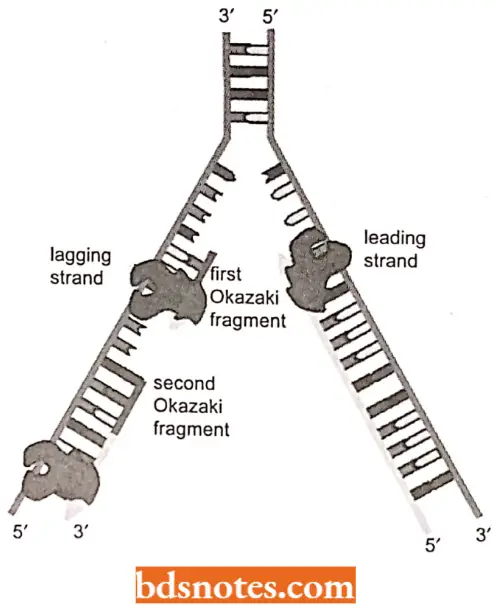
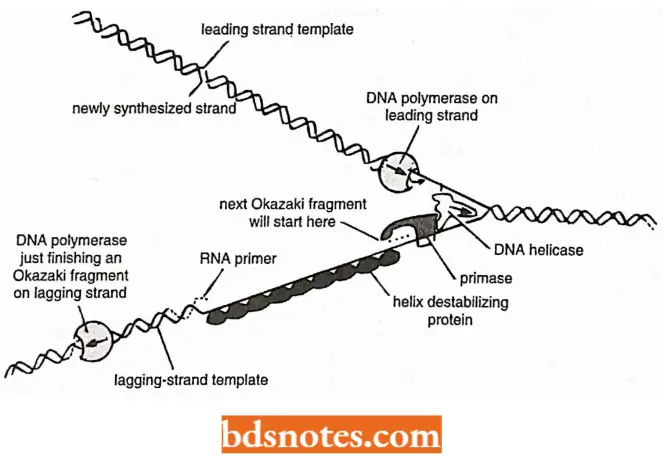
Semidiscontinuous DNA Replication: Various experimental evidences have suggested that DNA synthesis is continuous on one strand (3′ to 5′ strand), called leading strand and discontinuous on the other strand (5′ to 3′ strand), called lagging strand.
- Since DNA synthesis always proceeds in 5r to 3′ direction, so. on the lagging strand, synthesis takes place discontinuous!}’ in pieces, called Okazaki fragments (alter the name of discoverer R. Okazaki.
- 196S). Okazaki fragments average about 1,500 nucleotides in prokaryotes and 150 in eukaryotes (see Tamarin, 2002).
- Later on. these pieces arc fused with the help of ligase enzyme to form an intact lagging strand.
- Such a DNA replication, where the leading strand is synthesized continuously and the lagging strand is synthesized discontinuously, is called semidiscontinuous replication.
- Once initiated, continuous DNA replication can proceed indefinitely. DNA polymerase 111 on the leading – strand template has what is called processivity:
- once it is attaches, it does not release until the entire strand is replicated. Discontinuous replication, however, requires the repetition of four steps: primer synthesis, elongation, primer removal with gap tilling and ligation.
Unidirectional and Bidirectional DNA Replication: Regarding the direction of replication, DNA replication may be of the following two types:
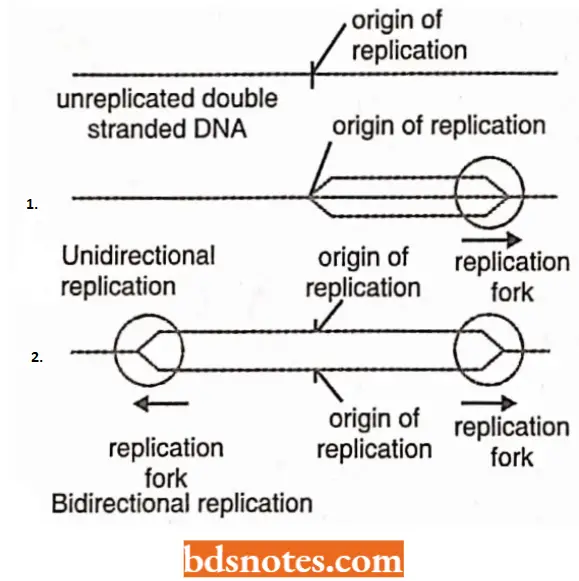
Bidirectional replication: All known DNA molecules, with only few exceptions, replicate as circles (or bubbles/eyes) and, hence, initiate within the helix.
- In the electron microscope, eukaryotic chromosomes are found to contain multiple expanding replication eyes, in contrast to a single eye of the prokaryotic DNA.
- Further DNA synthesis within a given replication unit eye is initiated some where at or near the midpoint of the unit at a site termed as the origin (O); prokaryotes contain a solitary origin, while eukaryotes have multiple (up to several thousands) origins for DNA replication.
- Both ends of the eye are moving and serve as replication forks.
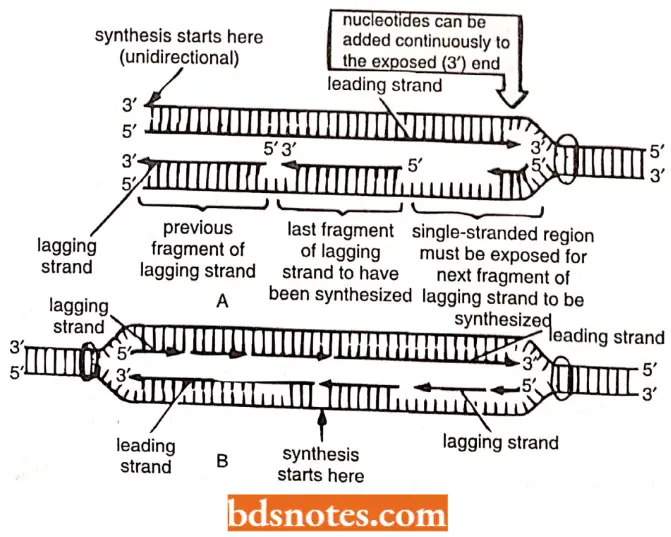
- Two replicating forks are then believed to travel in opposite directions until they reach either end of that unit, the two end points being called termini(T).
- A given replication unit may or may not undergo bidirectional synthesis in synchrony with continuous units.
- In either case the newly replicated strands in adjacent units will eventually meet. These strands are then linked, perhaps, by a DNA ligase to form long, continuous daughter DNA strands.
Unidirectional replication: In case of this type of DNA replication, one of two ends of the replication eye remains stationary and the other end serves as the replication fork and moves with replication.
An example of unidirectional replication is the replication of mitochondrial DNA (mt DNA) by D-loop (or displacement loop) in vertebrates.
Enzymes And Proteins Of DNA Metabolism
Different prokaryotic and eukaryotic cells have been found to contain three kinds of nuclear enzymes or enzymatic activities that act on DNA, namely nucleases, polymerase and ligases.
Enzymes involved in DNA Replication
- Nuclease enzymes: The nuclease enzymes act to hydrolyze or break down a polynucleotide chain into its component nucleotides. A polynucleotide is held together by 3′, 5′ phosphodiester bonds and a nuclease enzyme will attack either the 3′ or the 5′ end of this linkage. The nuclease enzymes may be of the following two kinds.
- Exonuclease enzymes: A nuclease enzyme which begins its attack from a free end of a polynucleotide is called exonuclease. Therefore, depending on the specificity of the enzyme, an exonuclease will either begin at a free 3′-OH end of a polynucleotide and progressively cleave the bonds on the 3r-OH side of the phosphodiester backbone or it will begin at a free 5′-P end and digest the polynucleotide in a 5’→ 3′ direction.
- In both cases the enzyme travels along the chain in a stepwise manner, liberating single nucleoside monophosphate molecules and eventually digesting entire polymer.
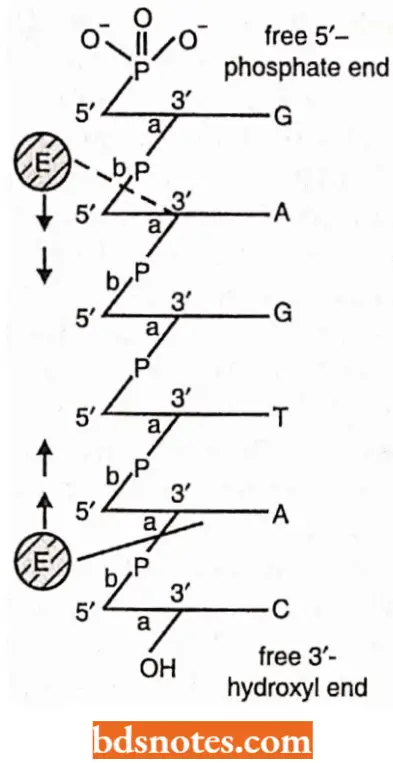
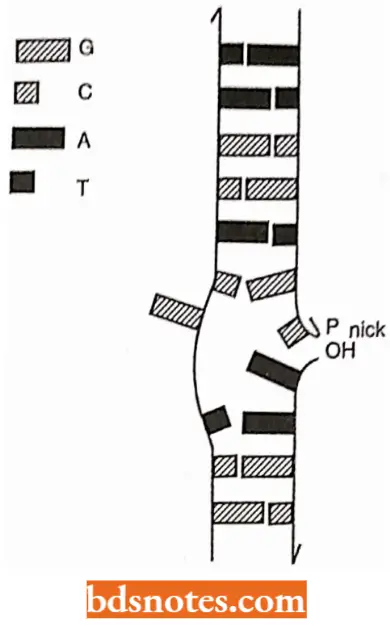
- Endonuclease enzyme: Endonuclease enzyme also attacks one of the two sides of phosphodiester linkages, but they react with those bonds that occur within the interior of a polynucleotide chain.
- If the polynucleotide chain is single-stranded (for example., viral DNA), such an attack will obviously cut the chain into two pieces. If, however, the polynucleotide strand is a member of a DNA helix (for example., prokaryotic and eukaryotic DNA), a single endonucleolytic cut will create a nick in the helix; the helix remains in one piece but it now possesses a gap that contains two free ends, which can serve as substrates for exonucleases.
- A nicked double helix suffers a localized disruption of its secondary structure. The molecules becomes free to bend or rotate around its intact strand and the two broken ends are free to dangle.
- The increased molecular motion in the region of the nick will very likely interrupt hydrogen bonding between bases in the vicinity of the nick, thus, effecting a limited “untravelling” of the helix.
Polymerase or replicase enzymes: A polymerase enzyme catalyses the formation of a polymer and cellular polymerase enzymes of genetic interest are those that bring about the synthsis of one polynucleotide chain that is a copy of another.
- A polymerase enzyme is called replicase enzyme when the copy of polynucleotide chain so produced is inherited by daughter cells or viruses, that is, when the enzyme brings about chromosome replication (see Goodenough and Levine, 1974).
In vitro DNA polymerization: To understand the mechanism of DNA-replication inside the living cell (in vivo), molecular biologists tried in vitro polymerization of DNA and found that in addition to DNA polymerase enzyme, three classes of organic molecules are essential for an in vivo reaction.
- The first are deoxynucleoside triphosphate. These are familiar deoxynucleotide monophosphates (i.e., dAMP,dCAMP,dGMP,dTMP) with two additional phosphate groups attached to the initial or α- phosphate group (i.e., dATP, dCTP, dGTP and dTTP).
- The second class of essential molecules for many DNA polymerase enzymes are polynucleotide chains with free 3′-OH ends (often called primer strands), meaning that they cannot initiate the de novo synthesis of a new strand.
- The third class of essential molecules for DNA polymerase enzymes arc template strands. All biologically important DNA polymerases possess a critical property:
- They will add nucleotides to a primer strand only in response to the base sequence found on a second template strand.
- Just as Watson and Crick suggested in their original model of DNA replication, the polymerase enzymes observe the rule of complementary base pairing.
- Thus, if a free 3’-OH group on a primer strand lies opposite a thymine on a template strand, a polymerase enzyme will add only an adenine group to the primer, even when dCTP, dTTP, dGTP are also present in the reaction mixture.
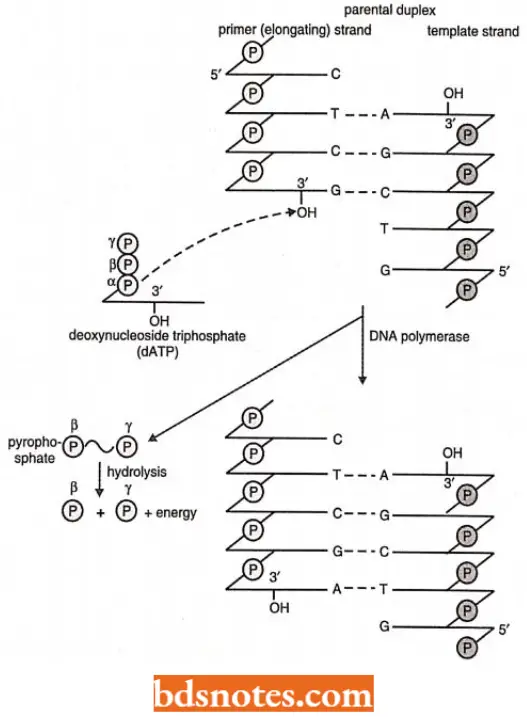
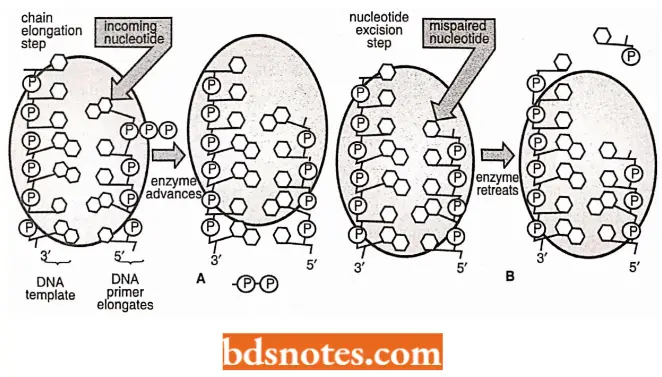
- Once the appropriate molecules are present and the appropriate ionic conditions are maintained, in vitro polymerization reaction summarized in will occur.
- The α-phosphate of a nucleoside triphosphate molecule forms a 3′, 5′ phosphodiester bond with a free 3′-OH in the growing polynucleotide chain, and a molecule of pyrophosphate (P ~ P) is simultaneously released.
- Pyrophosphate containes a “high-energy” or “ ~ “ bond, meaning that when the released pyrophosphate is hydrolyzed into two phosphate molecules, energy is liberated which drives the polymerization process forward.
- The resultant polymerization will always proceed in a net 5’→ 3′ direction, meaning that the nucleotide at the 3′ end is always the most recently added to the chain.
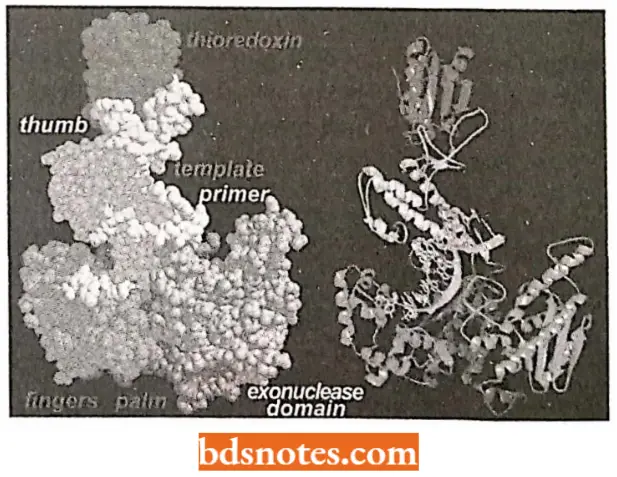
- Prokaryotic DNA polymerases. Three different DNA polymerases are known in E.coli and other prokaryotes, of which DNA polymerase 1 and 2 are meant for DNA repair and DNA polymerase III is meant for actual DNA replication.
DNA polymerase 1: This enzyme was isolated around 1960 by Arthur Kornberg and was the first enzyme suggested to be involved in DNA replication.
- It is also called Kornberg enzyme. DNA polymerase 1 enzyme is now considered to be a DNA repair enzyme rather than a replication enzyme.
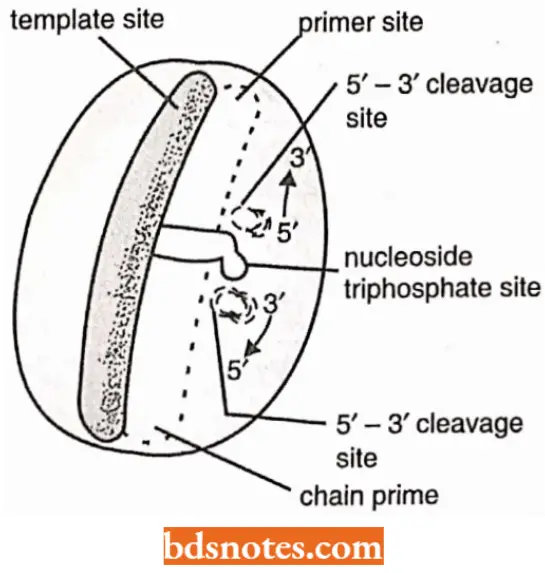
- This enzyme is known to have five active sites, namely template site, primer site, 5′ → 3′ cleavage or exonuclease site, nucleoside triphosphate site and 3’ → 5’ cleavage site (or 3′ → 5′ cxonuclcase site).
- DNA polymerase I is mainly involved in removing RNA primers from okazaki or precursor fragments and filling the resultant gaps due to its 5′ → 3′ polymerizing capacity.
- DNA polymerase 1 enzyme can also remove thymine dimers produced due to UV – irradiation and fill the gap due to excision.
- Both polymerization (= chain elongation) and exonuclease activity of DNA polymerase. This is called proof reading or editing function of this enzyme.
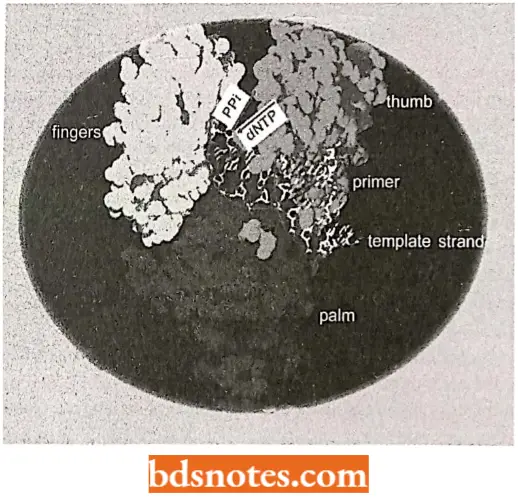
- Klcnow frngment/polymerasc. T. Stcitz and coworkers have done much X-ray crystallography work that has given us a good look at the structure of a DNA polymerase.
- In fact, most of their work has been done on a fragment of DNA polymerase I called Klenow fragment.
- Klenow fragment/polymerase was initially prepared by cutting the natural E.coli DNA polymerase I enzyme into two segments with a protease.
- One of these segments retained the polymerase and 3′ → 5′ exonuclease activity, but lacked the 5′ → 3′ exonuclease.
- Klcnow enzyme is shaped like a cupped right hand with enzymatic activity taking place in two places, separated by a distance of about two or three nucleotides.
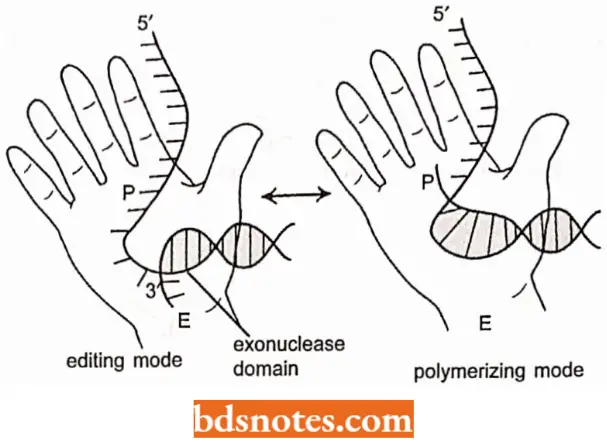
It is proposed that when the polymerization site senses a mismatch, the DNA is moved so that the 3′ end enters the exonuclease site, where the incorrect nucleotide residue is then cleaved. Polymerization then continue.
DNA Polymerase – 2: This enzyme resembles DNA polymerase-1 in its activity, but is a DNA repair enzyme. It brings about the growth in 5′ → 3′ direction, using free 3′-OH groups.
DNA polymerase-3: DNA polymerase-3 or Pol 3 enzyme plays an essential role in DNA replication. It is a multimeric enzyme or holoenzyme having ten subunits such as alpha (α), beta(β), epsilon (ε), theta (θ), tau (τ), gamma (γ), delta (δ), delta dash (δ’), chi (Χ) and psi(ψ).
- All these ten subunits are needed for DNA replication in vitro; however, all having different functions. For example, three of the subunits a, e and 0 form the polymerization core, with both 5′ →3′ polymerase activity and 3′ → 5′ exonuclease activity.
- One subunit, the P subunit, is a “processivity clamp”. As a dimer (two identical copies attached head to tail), the protein forms “doughnut” around the DNA so it can move freely on the DNA.
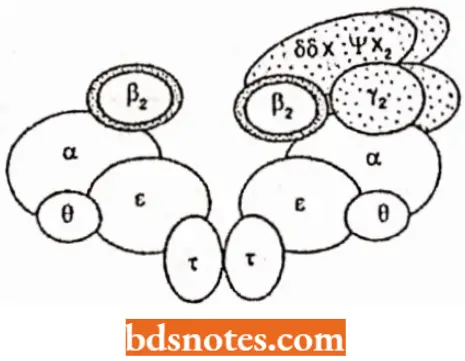
- subassembly without γ complex Is highly processive and probably functions In leading strand synthesis
- subassembly with γ complexcan dissociate fromtemplateand probably functions in lagging strand synthesis
- When it is attached to the core enzyme, the polymerase is held tightly to DNA and shows high proeessivity: the leading strand is usually synthesized entirely without the enzyme leaving the template.
- The remaining subunits are involved in proeessivity control and replisome formation. They allow polymerase to move ot V and on DNA o flagging-strand template as Okazaki fragments are completed (a process known as polymerase cycling).
Comparison Of Different Characters Of Three Types Of Dna Polymerases Of E.coli:
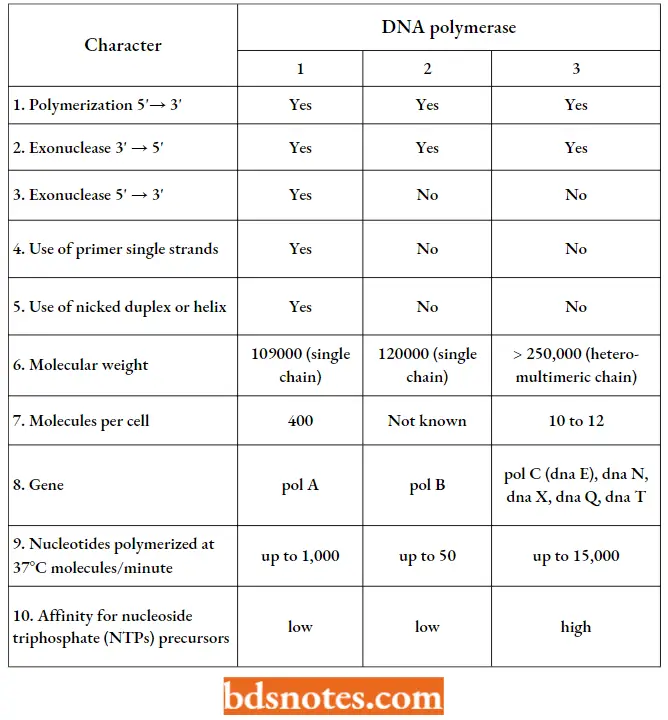
Eukaryotic DNA polymerases: Eukaryotes (for example., yeast, rat liver, human tumour cells) are found to contain the following five types of DNA polymerases:
- DNA polymerase α (=alpha): This relatively high molecular weight enzyme is also called cytoplasmic polymerase or large polymerase. It is found in nucleus and cytoplasm.
- DNA polymerase β (=beta): This enzyme is also called nuclear polymerase or small polymerase and is found only in vertebrates.
- DNA polymerase γ (=gamma): This enzyme is called mitochondrial polymerase and is encoded in the nucleus.
- DNA polymerase δ (=delta): This enzyme is found in mammalian cells and is PCNA dependent for DNA-synthesis processivity (PCNA = proliferating cell nuclear antigen).
- DNA polymerase ε (=epsiIon): It was previously known as DNA polymerase II. This enzyme is PCNA independent and occurs in mammalian HeLa cells and budding yeast.
The large DNA polymerase a is the predominant DNA polymerase enzyme in eukaryotic cells and was believed for long time to be only enzyme involved in DNA replication.
- But now one more polymerase, namely DNA polymerase 8 is also found to be involved in eukaryotic DNA replication.
- DNA ligases. DNA ligase enzymes are capable of catalyzing phosphodiester bond formation between free 3-OH and free 5-P groups of a nick of DNA which is created by endonuclease enzyme, thereby restoring an intact DNA duplex. Many DNA ligases have already bee discovered.
Summary Of The Enzymes And Proteins Involved In Dna Replication In E.coli:
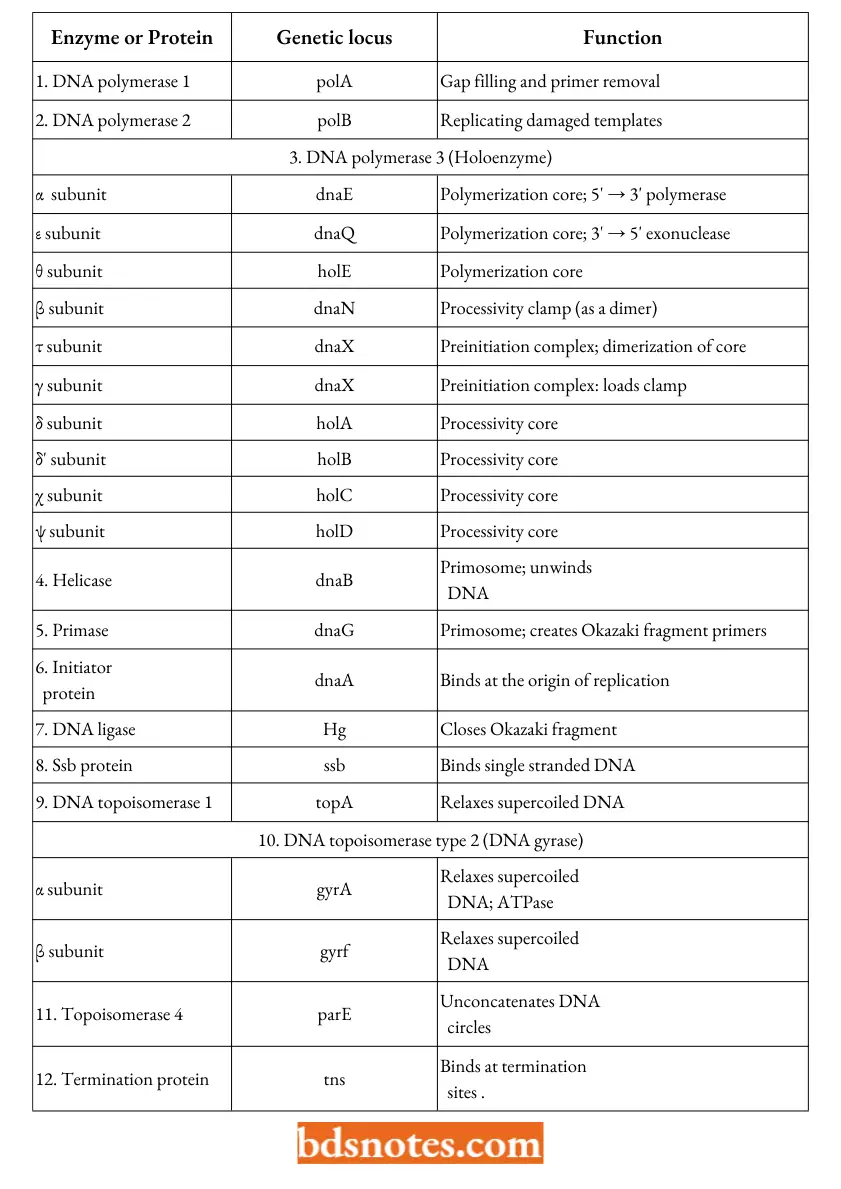
The ligase enzyme from E.coli requires the presence of oxidized nicotinamide adenine dinucleotide(NAD+) a cofactor, whereas the ligase enzyme specified by T4 bacteriophage requires ATP to bring about the joining reaction.
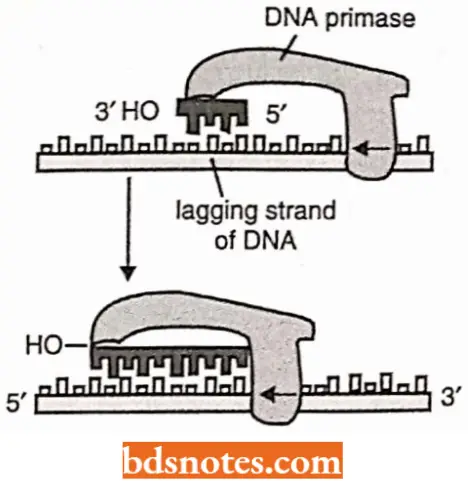
Roles Of DNA Primers In DNA Replication: No known DNA polymerase can initiate synthesis of DNA without the availability of a primer RNA strand.
- So, before actual DNA replication starts, short RNA oligonucleotide segments, called RNA primers or simply the primers, have to be synthesized by DNA primase enzyme utilizing ribonucleoside triphosphates.
- This RNA primer is synthesized by copying a particular base sequence from one DNA strand and differs from a typical RNA molecule in that after the synthesis the primer remains hydrogen-bonded to the DNA template(FreifeIder, 1985).
- The primers are about 10 nucleotides long in eukaryotes and they are made at intervals on the lagging strand where they are elongated by the DNA polymerase enzyme to begin each Okazaki fragment.
- These RNA primers are later excised and filled with DNA with the help of DNA repair system in eukaryotes (or DNA polymerase I in E.coli).
- In bacteria, two different enzymes are known to synthesize primer RNA oligonucleotides – RNA polymerase (on the leading strand) and DNA primase (on the lagging strand).
Replicons: DNA replication in prokaryotes and eukaryotes is attained in discrete units, called replicons. The number of replicons may vary in a genome from one in bacteria (.E.coli) and 500 in yeast to several thousands in plants and animals.
- For example, in the E.coli there is a single replication with the origin, identified as a genetic locus ori C (245 bp).
- The origin is A: T rich, a feature that is related to unwinding of DNA to initiate replication. In E.coli, there are also termination sites (terA-F), each consisting of ~23 bp.
- The process of termination of DNA replication requires the product of tus gene (Tus protein or TBP, i.e., ter binding protein) which recognizes ter or termination sites.
Prokaryotic And Eukaryotic Replicons:
![]()
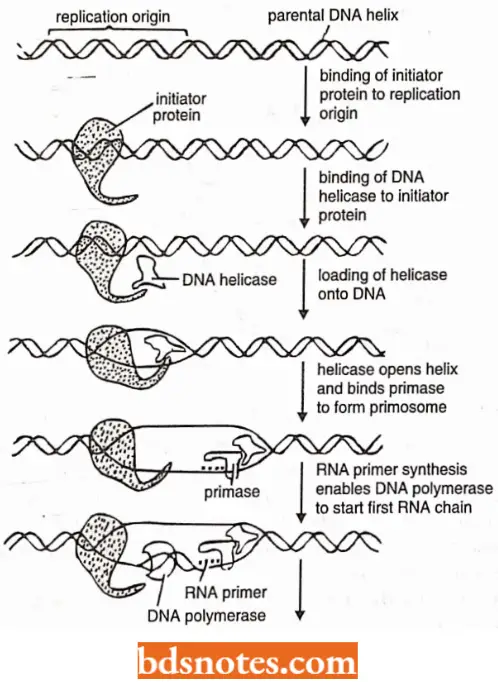
Proteins Involved In Opening Of DNA Helix: The following three types of proteins are needed to help the DNA double helix to open and to provide exposed DNA template for the DNA polymerase to copy:
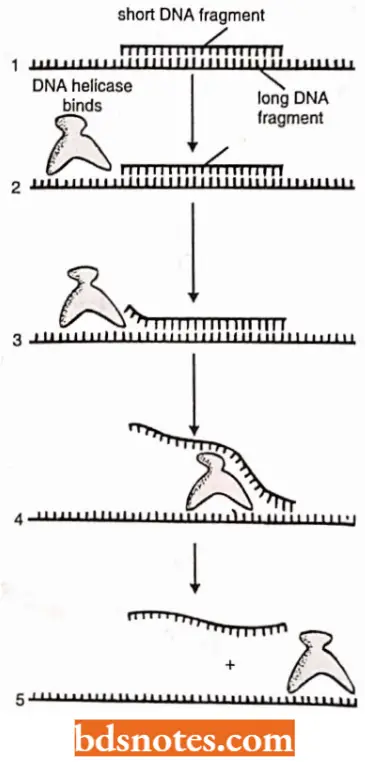
DNA helicases: DNA helicases are ATP-dependent upwinding enzymes which promote separation of the two parental strands and establish replication forks that will progressively move away from the origin.
- DNA helicase hydrolyze ATP when they are bound to single strands of DNA.
- Hydrolysis of ATP can change the shape of a protein molecule in a cyclic manner that allows the protein to perform mechanical work.
- DNA helicases utilize this principle to move rapidly along a DNA single strand; when they encounter a region of double helix, they continue to move along their strand, thereby unwinding the helix.
Unwinding of the template DNA helix at a replication fork could in principle be catalyzed by two DNA helicases, acting in concert, one running along the leading strand and the other along the lagging strand.
Helix-destabilizing strand, (also called single strand DNA-binding proteins or SSBPs): Behind the replication fork, the single DNA strands are prevented from rewinding about one another (or forming double-stranded hair-pin loops in each single strands) by the action of SSB proteins.
SSB proteins bind to exposed DNA strands without covering the bases, which, therefore, remain available for the templating process.
Topoisomerases (DNA gyrases): The action of a helicase introduces a positive supercoil into the duplex DNA ahead of the replication fork.
- Enzymes, called topoisomerases, relax the supercoil by attaching to the transiently supercoil duplex, nicking one of the strands and rotating it through the unbroken strand, the nick is then released.
- Thus, a DNA topoisomerase can be viewed as a “reversible nuclease” that adds itself covalently to a DNA phosphate, thereby breaking a phosphodiester bond in the DNA strand, because the covalent linkage that joins a topoisomerase to DNA phosphate retains the energy of the broken phosphodiester bond, the breakage reaction is reversible; resealing is rapid and does not require additional energy input.
- The rejoining mechanism is different from DNA ligase enzyme.
- One type of topoisomerase (i.e., topoisomerase I) causes a single-strand break or nick which allows the two sections of DNA helix on either side of nick to rotate freely relative to each other, using the phosphodiester bond in the strand opposite the nick as a swivel point.
- A second type of topoisomerase (i.e., topoisomerase 2) forms a covalent bond to both strands of DNA helix at the same time, making transient double-strand break in the helix.
Replisome and Primosome: N.K. Sinha and A. Kornberg have suggested that the DNA polymerases, RNA primases and helicases may be associated with one another to form a multienzyme complex – the replisome that carries out the synthesis of leading and lagging strands in a coordinated fashion.
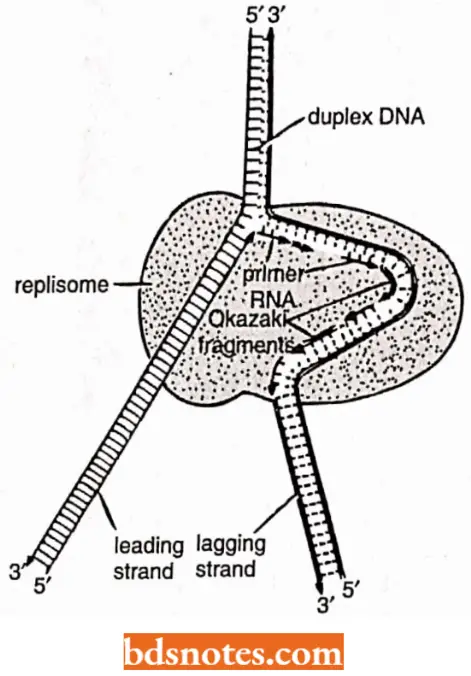
Such a complex would be highly processive and assure rapid replication of the DNA Likewise, the proteins at a replication fork cooperate to form replication machine, i.e., the primase molecule is linked directly to the helicase to form a unit on the lagging strand called a primosome which moves with the fork, synthesizing RNA primers as it moves.
Mechanism Of Dna Replication In Prokaryotes
In vitro DNA replication has been extensively studied in E.coli and in the phages and plasmids of E.coli. In E.coli, the process of DNA replication involves the following three steps.
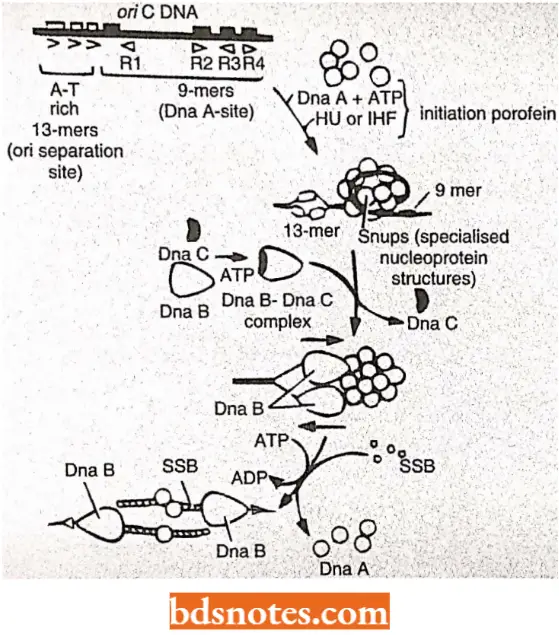
Initiation of DNA replication: This process comprises three steps:
- Recognition of the origin (O),
- Opening of DNA duplex to generate a region of single stranded DNA, and
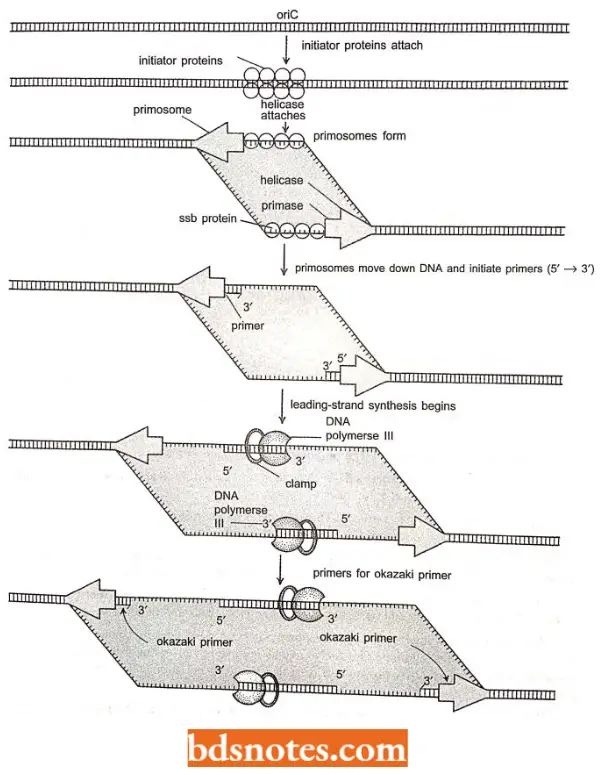
- Capture of Dna B protein (i.e., 5’→ 3′ helicase; also acts as the activator of primase). Thus, Dna- A (or initiator protein)-ATP complex binds at 9 bp inverted repeat regions(R1, R2, R3, R4) of ori C of E.coli and promotes opening of the DNA duplex in a region of three direct repeats of 13-bp sequence (called 13-mers).
The opening occurs from right 13-mer leftwards and requires negatively supercoiled DNA and HU or IHF initiator proteins.
- Dna B (=helicase) is transferred to exposed single stranded DNAand causes unwinding of the DNA in the presence of ATP, SSB protein and DNA gyrase (a topoisomerase).
- This results in unwinding of DNA duplex and the replication from ori proceeds in both directions (bidirectional); SSB binding occurs on single stranded regions and two Dna B complexes (=primosomes) are loaded one on each strand.
Elongation of DNA chain: This step requires the presence of the following enzymes and factors:
- Dna B or helicase (also called mobile promoter);
- Primase (Dna G);
- DNA polymerase holoenzyme (or DNA pol 3 HE);
- SSB proteins;
- RNAase H which removes RNA primers;
- DNA polymerase I which is used for filling the gap created due to RNA primers and
- DNA ligase (which converts primerless Okazaki fragments into continuous strand).
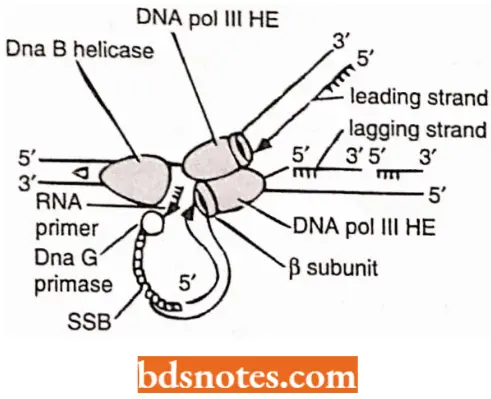
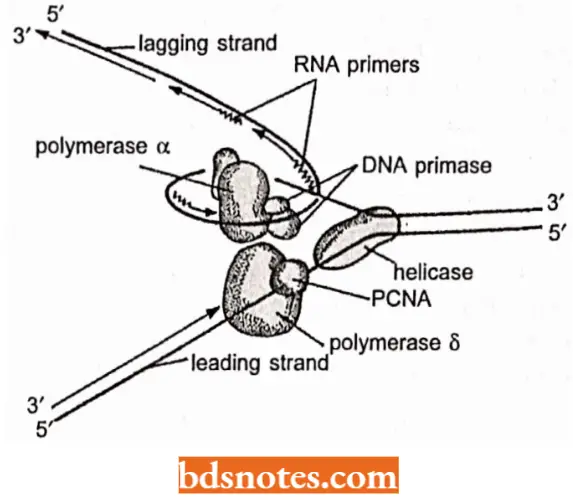
During initiation to elongation transition, the following events occur:
- As helicase (or Dna B) travels in 5′ → 3’ direction, it generates a replication fork by opening the DNA duplex.
- The DNA strand having helicase becomes the lagging strand. DNA primase associates with Dna B helicase, forming the primosome which synthesizes multiple primers for lagging strand and single RNA primer for the leading strand.
- For the synthesis of lagging strand, the DNA pol 3 HE has to work on the same strand to which Dna B helicase is bound, but it travels in opposite direction.
- Dna B helicase, Dna G primase and DNA pol 3 HE work together in strand elongation. Helicase and DNA polymerase assembly remains processive, i.e., they remain tightly bound to the fork and stay bound through the reaction.
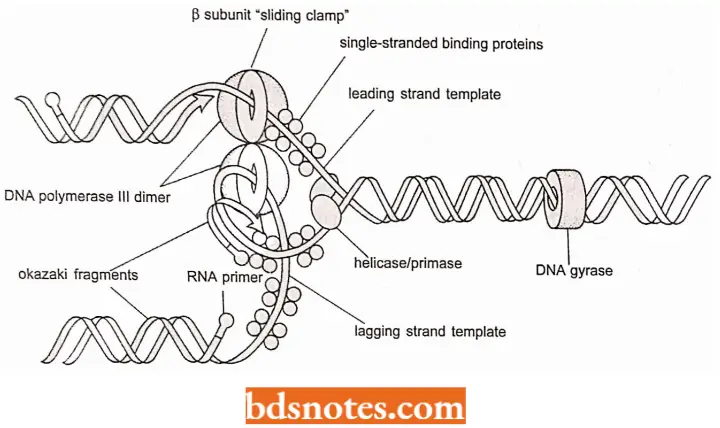
Synthesis (=elongation) of lagging and leading strands takes place by somewhat different methods; it is far more complex for lagging strand than for the leading strand:
Discontinuous synthesis on lagging strand: Primase is taken up from solution and is activated by helicase (Dna B) to synthesize a RNA primer (10 to 20nt or nucleotides long) on the lagging strand.
- The RNA primers are recognized by DNA pol 3 HE on the lagging strand and arc utilized for synthesis of precursor or Okazaki fragments.
- In fact, each new RNA primer is recognized by the gamma (γ) subunit of DNA pol 3 HE and loaded with a subunit of the same polymerase.
- This preloaded β subunit may then capture the core of DNA poly 3 HE when it becomes available after finishing its synthetic job on the preceding Okazaki fragments.
- On completion of the Okazaki fragments, the RNA primers are excised by DNA polymerase 1, which then fills the resulting gaps with DNA.
- After DNA polymerase 1 adds the final deoxyribonucleotides in the gap left by the excised primer, the enzyme DNA ligase forms the phosphodiester bond that links the free 3′ end of the primer replacement to the 5′ end of the Okazaki fragment.
Continuous Synthesis On Leading Strand
- In bidirectional DNA replication, the leading strand is primed once on each of the parental strands.
- The RNA primer of the leading strand is synthesized by RNA polymerase enzyme.
- DNA pol 3 HE causes elongation of the leading strand and finally DNA pol 1 and ligase enzymes give final touch to the leading strand as in case of the lagging strand.
Dna Replication In Eukaryotes
Eukaryotic DNA replication requires two different DNA polymerase enzymes, namely DNA polymerase a and DNA polymerase δ.
- DNA polymerase δ synthesizes the DNA on the leading strand (continuous synthesis), whereas DNA polymerase a synthesizes the DNA on the lagging strand (discontinuous DNA synthesis).
Besides these two enzymes, six more factors are involved in eukaryotic DNA replication:
- T antigen;
- Replication factor A or RF-A (also called RP- A or eukaryotic SSB);
- Topoisomerase 1;
- Topoisomerase 2;
- Proliferating – cell nuclear antigen (PCNA), also called cyclin), and (a) replication factor C or RF-C.
The process of eukaryotic DNA replication involves the following steps:
- Before the onset of DNA synthesis, there is a presynthetic stage of 8 – 10 minutes duration for the formation of unwound DNA complex. This step needs only three purified proteins, namely T antigen (T-ag or tumour antigen), RF-A and topoisomerase 1 and 2.
- The T-antigen, using its DNA-binding domain, forms a multisubunit complex with site 1 and site 2 in the presence of ATP and caused local unwinding.
- More extensive duplex unwinding occurs due to association of RF-A and a topoisomerase with the help of DNA helicase. Topoisomerases help in unwinding of DNA by altering topology of DNA at the replication fork.
- RF-A or SSB proteins bind to unwound single stranded DNA.
- The primer RNA synthesis is performed by primase which is tightly associated with DNA polymerase α.
- DNA polymerase a helps in synthesis of an Okazaki fragment in 5′ to 3′ direction. In eukaryotes, Okazaki fragments are much smaller – only about 135 bases long, about the size of the DNA on a nucleosome.
- Replication factor C (or RF-C) and PCNA (cyclin) help in switching of DNA polymerases so that pol α is replaced by pol δ which then continuously synthesized DNA on the leading strand.
- Another Okazaki fragment is then synthesized from the replication fork on the lagging strand by pol α-primase complex and this step is repeated again and again, till the entire DNA molecule is covered.
- The RNA primers are removed and the gaps are filled as in prokaryotic DNA replication.
Recently, role of DNA polymerase e in DNA replication has been stressed upon, so that three DNA polymerases (a, 5 and s) are now known to be involved in eukaryotic DNA replication.
Sugino and coworkers have proposed that DNA polymerase a might function at both the leading and lagging strands (since polymerase a has primase activity), whereas polymerase e and polymerase δ are involved in elongation of the leading and lagging-strands respectively.
Models Of DNA Replication
The following three models have been proposed for DNA replication in different organisms:
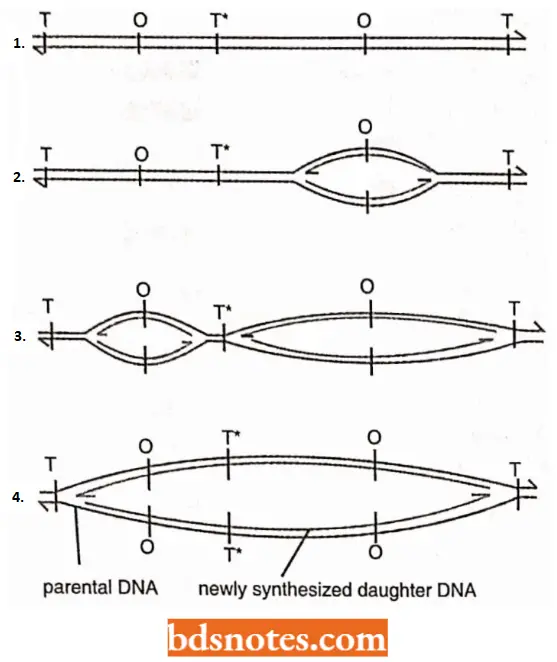
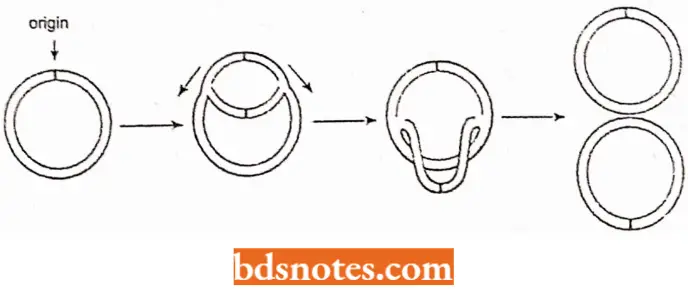
Replication Fork Model: It occurs both in linear and circular DNA molecules and involves the formation of replication forks which either move in one direction in unidirectional replication or both directions in bidirectional replication.
In E.coli, replication of circular DNA is of bidirectional type involving intermediate theta (θ) type structures.
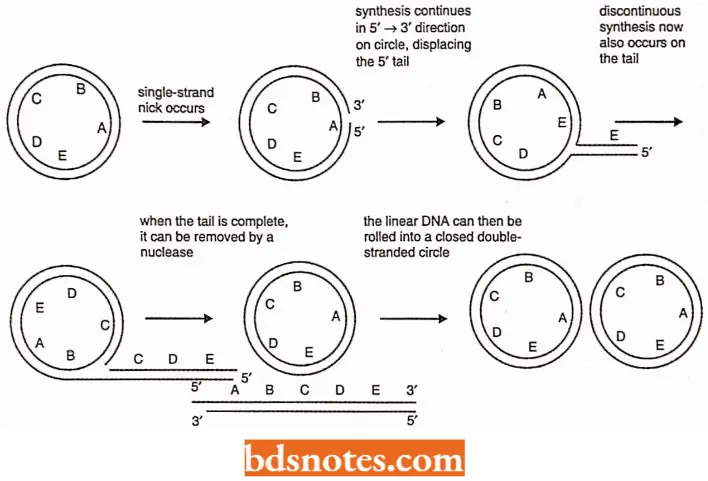
Rolling Circle Model (Rolling Circle Replication): In the rolling circle mode of DNA replication, a nick (a break in one of the phosphodiester bonds) is made in one of the strands of the circular DNA and this nick has 3′ – OH and 5′ – P termini.
- Due to a helicase and Ssb protein α, Y-fork is generated. Ultimate result is replication of a circle and a tail. This form of replication occurs in F plasmid or E.coli Hfr chromosome during conjugation.
- The F+ or Hfr bacterial cell retains the circular daughter while passing the linear tail into the F– cell, where replication of the tail takes place.
- Several phages (including 1 phage) also use this DNA replication method, filling their heads (protein coats) with linear DNA replicated from the circular molecule.
- Rolling circle model is also called sigma (s) replication and it is an efficient mechanism for rapid synthesis of multiple copies of a circular genome (Novick, 1998).
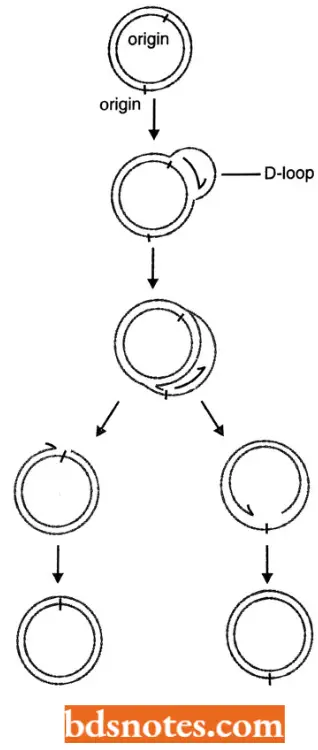
D-loop Model (Displacement Replication): Chloroplasts and mitochondria (in eukaryotic cells) have their own circular DNA molecules that replicate by a slightly different mechanism.
- The origin of replication h at a different point on each of the tv/o parental template strand, Replication begins on one strand, displacing the other while forming a displacement loop or 1) loop structure.
- Implication continues until the process passes the origin of replication on the other strand, Replication then initiates on the second strand, in the opposite direction, Y junction for replication fork) replication, also occurs in mitochondrial DNA under some grov/lh condition.
Termination Of Replication
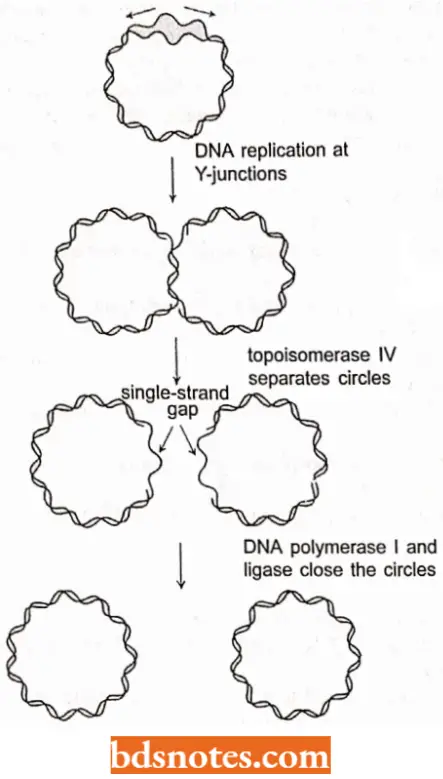
The lermiriafion of the replication of a circular chromosome DMA (of bacteria, mitochondria and chloroplasls) presents no major topological problems, Al the end of the theta structure replication, both Y- junctions have proceeded around the molecule.
- The region of termination on the E.coli chromosome, the terminus (Ter) is 180 degrees from OriC on the circular chromosome, between minutes 28 and 36.
- There are six terminator sites; three arrest the Y-junction from the left, and three arrest the one from the right when bound by a termination protein, the product of the tus gene. (Tus stands for terminus utilization substance; each ter site is about twenty base pairs).
- One interesting aspect of the termination of li.coli DNA replication is that the cells arc viable even if the whole terminator region is deleted.
- There are fewer viable cells and some growth problems, but in general, E.coli can successfully terminate DNA replication even without formal termination sites, A topoisomerasc, (lopoisomerasc IV) then releases the two circles and DNA polymerase I and ligase enzymes close them up.
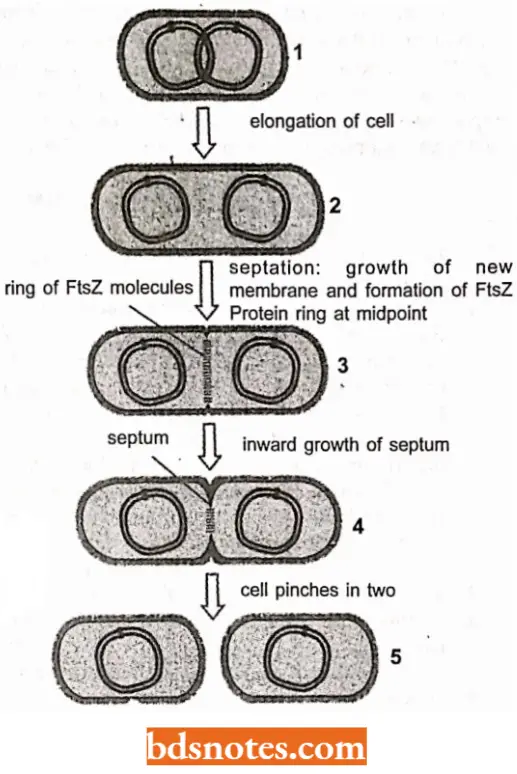
DNA Partitioning in E.coli: Until very recently, geneticists believed that the partitioning of the E.coli chromosome was a passive process, unlike that in eukaryotes.
- Now, however, wc know that E.coli DNA partitioning is indeed a very complex process.
- When DNA replication begins, the newly replicated origins of replication are segregated to opposite ends ofthe bacterial cell, acting as centromeres do.
- A ring of FtsZ protein, the products ftsZ gene, form a ring at the middle of the already elongated cell and begins to create septum that will divide the already elongated cell into two daughter cells.
DNA Replication Questions And Answers
Question 1. What is meant by the terms primer and template?
Answer:
Primer means nucleotide bound to DNA and having a 3′ -OH group; template means a polynucleotide strand whose base sequence can be copied.
Question 2. What is the role of RNA in DNA replication?
Answer: RNA serves as a primer.
Question 3. Distinguish the roles of helicases and SSB proteins in DNA replication.
Answer:
A helicase unwinds a helix. The SSB proteins prevent the helix from rewinding and prevent intramolecular base pairing from occurring.
Question 4. If one strand of DNA is found to have the sequence5’AACGTACTGC3’, what is the sequence of nucleotides on the 3’, 5’ strand?
Answer: 3’TTGCATGACG5′ because of pairing qualities of deoxyribonucleotides.
Question 5. For a molecule of n-deoxyribonucleotide pairs, give a mathematical expression that can be used to calculate the number of possible sequences of those nucleotide pairs if only the “usual” bases are present.
Answer: 4n
Question 6. Develop a formula for determining the length in micrometers of a DNA molecule whose number of deoxyribonucleotide pairs is known.
Answer:
L μm- 3.4 x 1 0-4P, where L\un represents the length in micrometers, and represents the number of nucleotide pairs.
Question 7. Phage T2 DNA is estimated to consist of about 200,000 deoxyribonucleotide pairs. What is the length in micrometers of its DNA complement?
Answer: 68 μ m.
DNA Replication Multiple Choice Questions And Answers
Question 1. DNA replication occurs in
- G, phase
- G2 phase
- S phase
- Interphase
Answer: 3. S phase
Question 2. Duplication of DNA is called
- Transduction
- Transcription
- Replication
- Translation
Answer: 3. Replication
Question 3. The process of DNA replication is
- Dispersive
- Conservative
- Semi-conservative
- Non-conservative
Answer: 3. Semi-conservative
Question 4. Semiconservative DNA replication was first demonstrated by
- Taylor
- Watson and Crick
- Meselson and Stahl
- Khorana
Answer: 3. Meselson and Stahl
Question 5. Taylor demonstrated that DNA replication is semi-conservative in
- Vicia faba
- Mouse liver cells
- Peanut
- HeLa cells
Answer: 1. Vicia faba
Question 6. Isotopes used for proving semiconservative replication are
- N14 and N31
- N14 and C14
- H3and N15
- C14 and P31
Answer: 3. H3and N15
Question 7. The unwinding of DNA duplex is performed by an enzyme called
- Gyrase
- Lactase
- Maltase
- ligase
Answer: 1. Gyrase
Question 8. The enzyme that reduces the tension during the unwinding of DNA helix in front of the
replication form is
- Topoisomerase
- Heliease
- Ligase
- Polymerase
Answer: 1. Topoisomerase
Question 9. During DNA replication the strands separate by
- DNA polymerase
- Gyrase
- DNA heliease
- DNA primase
Answer: 3. DNA heliease
Question 10. During DNA replication, the term leading strand is applied to the one which replicates in
- 5′ – 3′ direction continuously
- 3′ – 5′ direction continuously
- 5′ – 3′ direction discontinuously
- 3′ – 5′ direction discontinuously
Answer: 1. 5′ – 3′ direction continuously
Question 11. Which of the following enzymes makes short RNA chains using a DNA template?
- DNA polymerase I
- DNA helicase
- DNA primase
- DNA gyrase
Answer: 3. DNA primase
Question 12. Okazaki fragments are formed in
- The splicing of mRNA
- The synthesis of the lagging strand of DNA
- The neurons of vertebrates
- The oocytes of amphibians
Answer: 2. The neurons of vertebrates
Question 13. The enzyme that removes the RNA primer after DNA replication is
- Primase
- DNA polymerase 1
- DNA polymerase 2
- ligase
Answer: 2. DNA polymerase 1
Question 14. The enzyme that joins DNA fragments is
- Restriction endonuclease
- Lipase
- Ligase
- Peroxidase
Answer: 3. Ligase

Leave a Reply